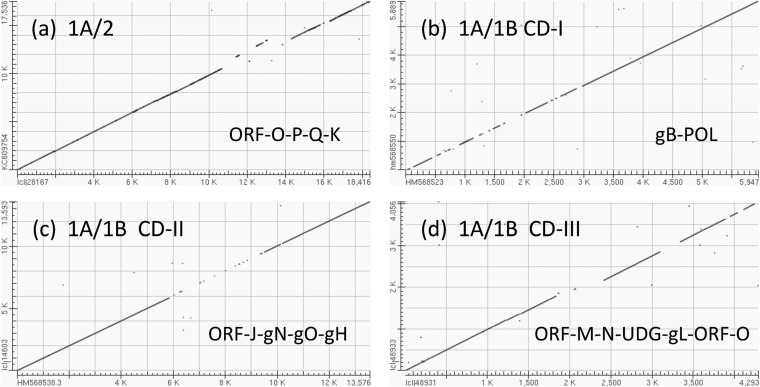
FIG 4.
Dot matrix alignments encompassing major areas of divergence for EEHV2 or EEHV1B versus EEHV1A. Examples of DNA level dot matrix alignments produced in BLAST2.2.22 as available online at NCBI. (a) Highly divergent ORF-O-P-K locus in EEHV2. The alignment from U74(PAF) to U86.5(ORF-L) was created by comparing EEHV2(NAP12) positions 1 to 17,536 from GenBank accession no. KC609754 with EEHV1A(NAP23, Kimba) positions 137,7000 to 156,117 from GenBank accession no. KC618527. Match-mismatch settings were 2,3, and gapcode was 2,2. The four mismatch gaps encompass parts of ORF-O, -P, -Q, and -K in EEHV1A, with the displaced segment resulting from the absence of ORF-Q in EEHV2. (b) CD-I locus in EEHV1B. Comparison of U39(gB) and U38(POL) using GenBank accession no. HM568523 (positions 1 to 5,947) from EEHV1A(NAP18, Kala) with GenBank accession no. HM568550 (positions 1 to 5,899) from EEHV1B(NAP19, Haji). Match-mismatch settings were 1,4, and gapcode was 5,2. (c) CD-II locus in EEHV1B. Comparison of U29(TRI1) to U51(vGPCR1) using GenBank accession no. HM568525 (positions 1 to 13,593) from EEHV1A(NAP18, Kala) with GenBank accession no. HM568538 (positions 1 to 13,593) from EEHV1B(NAP14, Kiba). Match-mismatch settings were 1,−3, and gapcode was 2,2. The central 3.2-kb gap represents part of U45.7(ORF-J), plus all of U46(gN), U47(gO), and U48(gH). (d) CD-III locus in EEHV1B (left-hand side [LHS]). Comparison of U77(HEL) to U85.5(ORF-O) using GenBank accession no. JX011082 (positions 223 to 4,279) from EEHV1B(NAP14, Kiba) with GenBank accession no. KC854707 (positions 1 to 4,293) from EEHV1A(NAP18, Kala). Match-mismatch settings were 1,−2, and gapcode was linear. The small displacement at the first gap represents a 220-bp deletion within the ORF-N(vCXCL1) gene of EEHV1B.