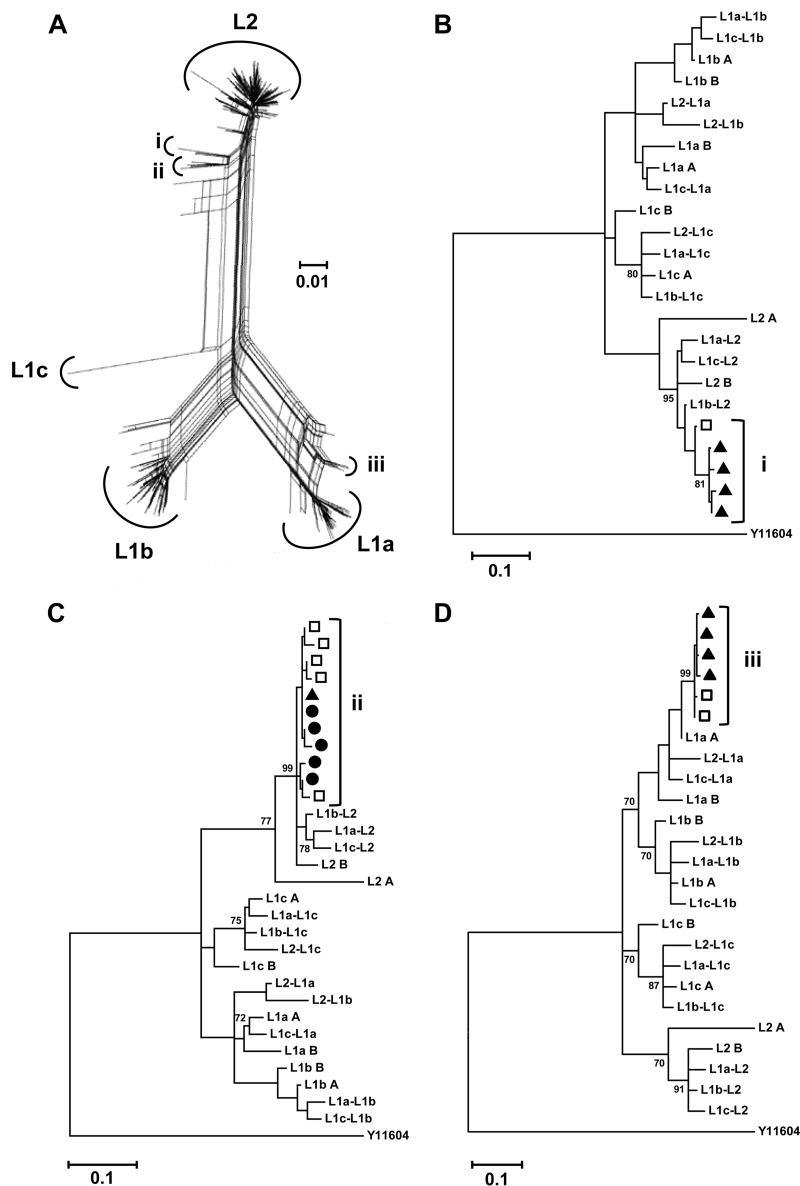
FIG 4.
Intersample maintenance of recombinants. (A) Composite phylogenetic network of the RL7-to-RL10 subset. Haplotype clusters displaying split decomposition signals are identified (i to iii). (B to D) Phylogenetic analysis of clusters i to iii, consensus sequences of the (sub)lineages, and interlineage recombinant sequences generated in silico using a rooted general time-reversible model (GTR + G + I). The data demonstrate intersample maintenance and evolution of recombinant haplotypes arguing in favor of the hypothesis of the recombinant sequences arising in situ within the host. Sequences are identified by time points as black circles (RL7), open squares (RL8), and black triangles (RL9). Sequences arising from RL10 did not feature in groups i to iii. A genotype 4a strain (Y11604) was used as the reference outgroup. Bootstrap values (of 1,000 resamplings) above 70 are shown. The genetic distance of 0.01 nucleotide substitutions per site is given by the scale bar.