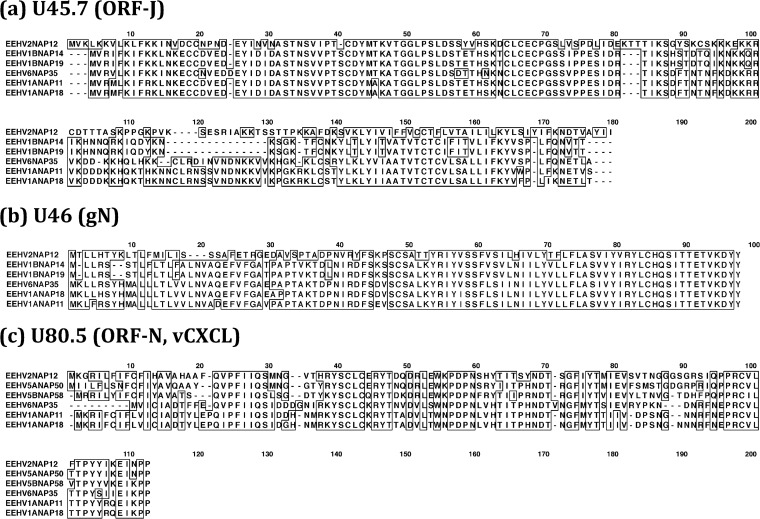
FIG 10.
Clustal alignments illustrating variability patterns among the intact ORF-J, glycoprotein N (gN), and ORF-N(vCXCL1) proteins of AT-rich branch EEHV genomes. Clustal-W presentations comparing the primary amino acid sequence alignments for all available predicted Proboscivirus versions of three small unique or poorly conserved proteins that do not have orthologues in other herpesviruses for comparison are shown. (a) U45.7(ORF-J), intact novel proteins. (Data are not yet available for ORF-J of EEHV5A or EEHV5B.) (b) U46(gN), glycoprotein N, intact proteins. (Data are not yet available for gN of EEHV5A or EEHV5B.) (c) U80.5(vCXCL1, ORF-N), possible inhibitory alpha-chemokine ligand. [The vCXCL1(ORF-N) gene is absent from EEHV1B.] Positions with 100% similarity are boxed.