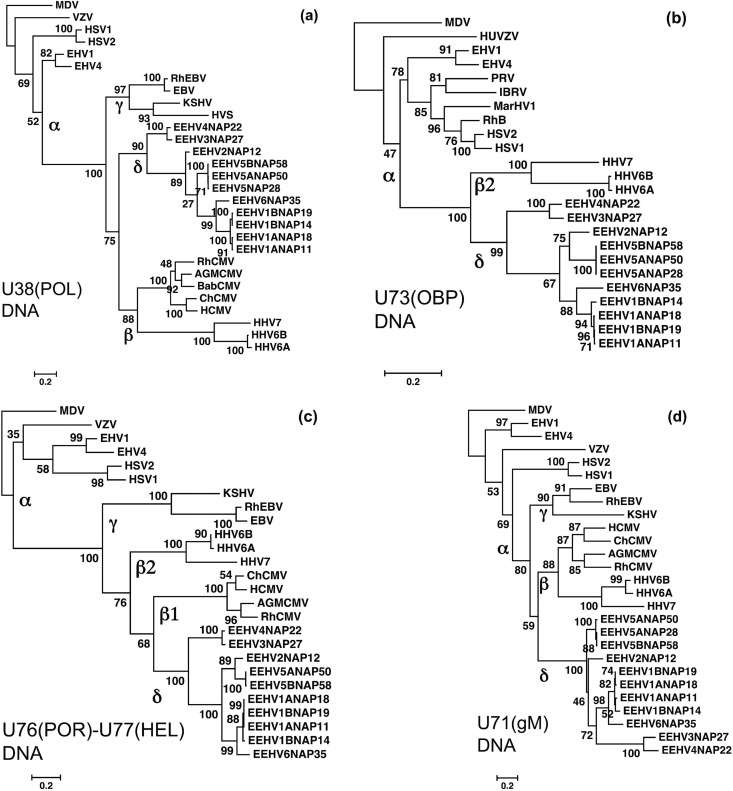
FIG 6.
Linear DNA level phylogenetic trees comparing four gene loci from EEHV3, EEHV4, EEHV5A, EEHV5B, and EEHV6 with their orthologues in EEHV1, EEHV2, and other key herpesviruses. The diagrams present linear Bayesian maximum likelihood phylogenetic dendrograms at the nucleotide level for a set of representative EEHV gene loci from the five EEHV3, EEHV4, EEHV5A, EEHV5B, and EEHV6 (proposed Deltaherpesvirinae [δ]) genomes determined here, together with orthologues from EEHV1A, EEHV1B, and EEHV2 (10). These are compared with matching segments of selected orthologous gene loci among representative herpesviruses from all three other mammalian subfamilies, Alphaherpesvirinae (α), Gammaherpesvirinae (γ), and Betaherpesvirinae (β). Alternative virus names and GenBank accession numbers for both the non-EEHV and EEHV genome file sources used are listed in supplemental material or in Table S1 in the supplemental material. Bootstrap values are shown as percentages at the nodes. The final DNA segment sizes were as follows for panels a to d: (a) U38(POL) locus, DNA polymerase, 927 bp; (b) U73(OBP) locus, origin binding protein, 576 bp; (c) U76(POR)-U77(HEL) locus, portal protein plus helicase subunit; (d) U71-U72(gM) locus, myristylated tegument protein plus glycoprotein M, 353 bp. All four panels use Marek's disease alphaherpesvirus (MDV) as an outgroup.