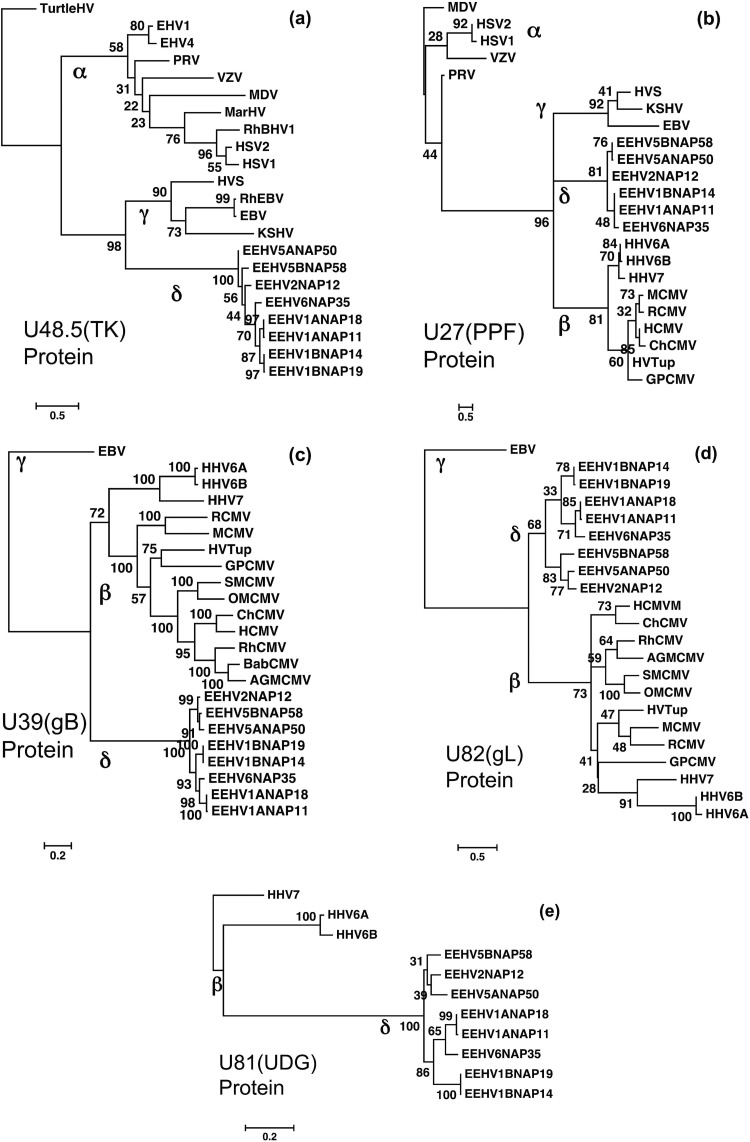
FIG 7.
Linear protein level phylogenetic trees comparing five protein segments from EEHV5 and EEHV6 with their orthologues in EEHV1, EEHV2, and other key herpesviruses. The diagrams present linear Bayesian maximum likelihood phylogenetic dendrograms at the amino acid level for a set of representative proteins from the prototype EEHV5A, EEHV5B, and EEHV6 genomes, as well as up to five other orthologues from EEHV1A, EEHV1B, and EEHV2 (proposed Deltaherpesvirinae [δ]). These are compared to matching segments of orthologous gene loci from selected herpesviruses representative of the Alphaherpesvirinae (α), Gammaherpesvirinae (γ), or Betaherpesvirinae (β) subfamilies. Bootstrap values are shown as percentages. The final protein segment sizes used were as follows for panels a to e: (a) U48.5(TK), thymidine kinase, 244 aa, with turtle herpesvirus (TurtleHV) used as the outgroup; (b) U27(PPF), polymerase processivity factor, 110 aa, with MDV used as the outgroup; (c) U39(gB), glycoprotein B, 727 aa, including only the Betaherpesvirinae as references with EBV used as the outgroup; (d) U82(gL), glycoprotein L, 85 aa, with only Betaherpesvirinae included as references and EBV used as the outgroup; (e) U81(UDG), uracil DNA glycosylase, 252 aa, with only Roseolovirus species used for reference and HHV7 used as the outgroup.