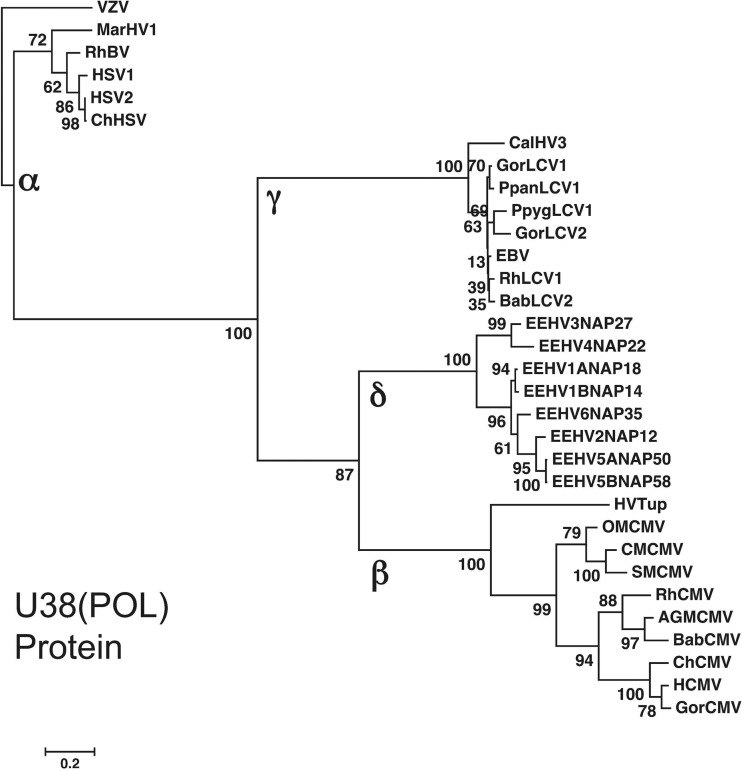
FIG 8.
Comparison of EEHV POL protein divergence levels with great ape plus New World and Old World primate versions within the alpha-, beta-, and gammaherpesvirus subfamilies. The diagram shows a linear distance-based Bayesian phylogenetic tree dendrogram at the amino acid level for each of the eight prototype EEHV genomes (proposed Deltaherpesvirinae [δ]) across the largest segment of U38(POL) in common (1,080 bp, Kimba equivalent coordinates 77,783 to 78,863). To allow direct comparisons of estimated ages of divergence (see the text), these are compared with matching segments of orthologous gene loci from selected primate herpesviruses representative of just the Simplexvirus (Alphaherpesvirinae [α]), Lymphocryptovirus genus (Gammaherpesvirinae [γ]), and Cytomegalovirus (Betaherpesvirinae [β]) genera. Human VZV (Varicellovirus genus) was used as the outgroup. The evolutionary history was inferred by using the maximum likelihood method based on the Kimura two-parameter model with initial trees for the heuristic model obtained by applying the neighbor-joining method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA5. Bootstrap values are shown as percentages.