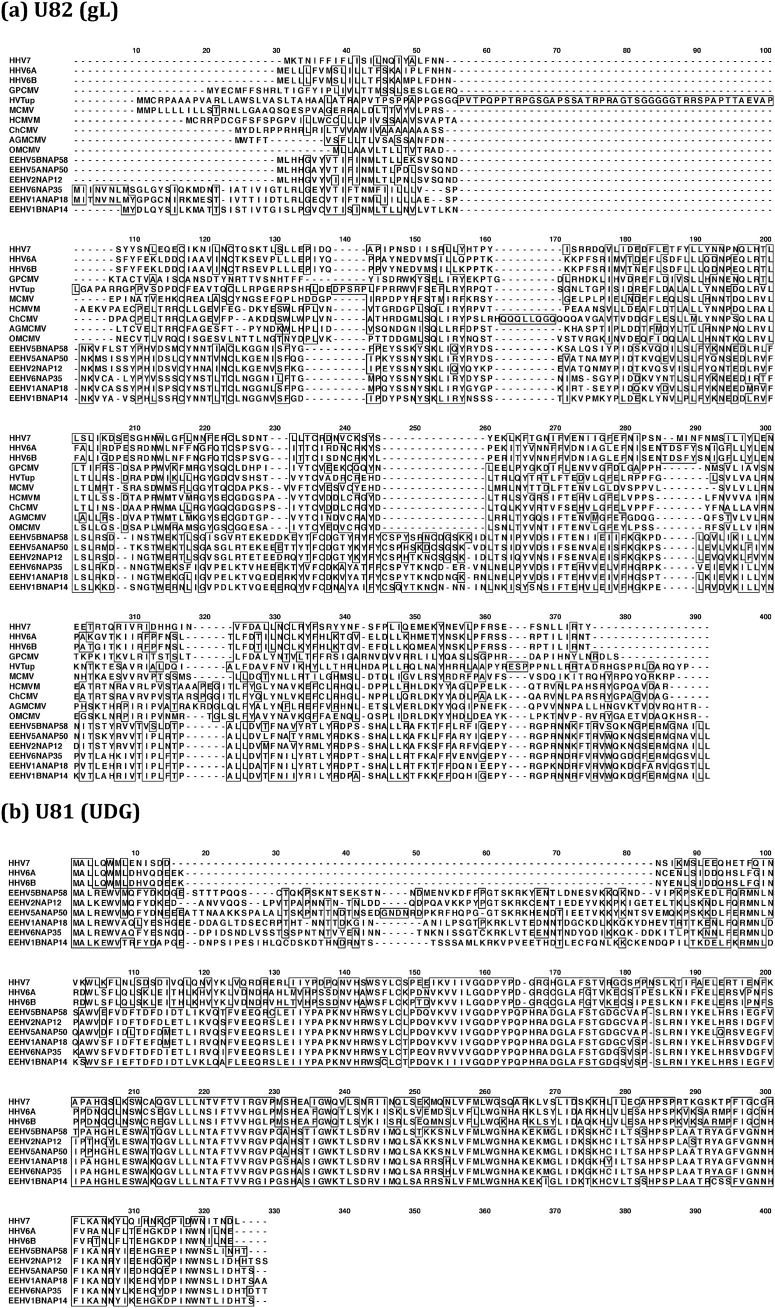
FIG 9.
Clustal alignment illustrating variability patterns among the intact glycoprotein L (gL) and uracil DNA glycosylase (UDG) proteins from distinct types of EEHV genomes. Clustal-W presentations comparing the predicted primary amino acid sequence alignments of the prototypes of all six available types and subtypes of AT-rich branch Proboscivirus genomes compared to their closest orthologues in either all Betaherpesvirinae or in just the Roseolovirus genus. GenBank accession numbers for the EEHV proteins and betaherpesvirus reference species used are given in the supplemental material. (a) U82(gL), glycoprotein L, intact proteins; (b) U81(UDG), uracil DNA glycosylase, intact proteins. Positions with 100% similarity are boxed. Gaps introduced to maximize alignment are indicated by hyphens.