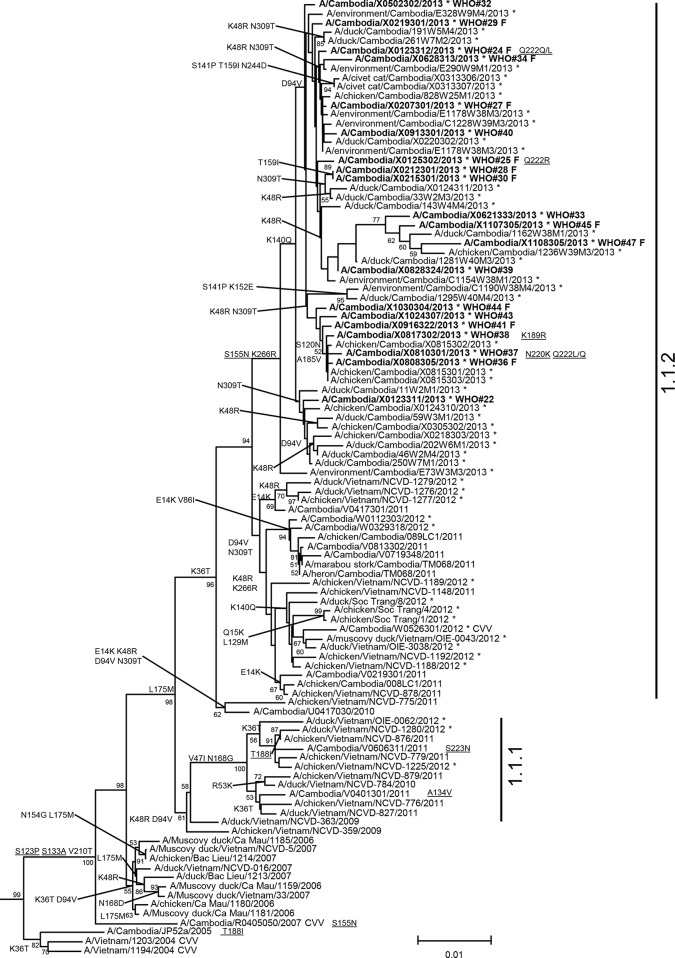
FIG 1.
Neighbor-joining phylogenetic tree of the HA genes of clade 1 highly pathogenic avian influenza A(H5N1) viruses constructed in MEGA5. The nearest reassortant WHO candidate vaccine viruses (CVV) for each group of clade 1 are denoted by CVV at the end of the strain name. Viruses collected in 2012-2013 are denoted with an asterisk. Sequences were aligned using MUSCLE, and amino acid differences at branch nodes indicate shared HA1 substitutions relative to the reference strain, A/Vietnam/1203/2004. Mutations to the right of a strain name indicate amino acid changes found only in that individual virus. Underlined amino acid substitutions indicate previously recognized molecular markers and/or markers of note as listed in the H5N1 Genetic Changes Inventory (24). Branches on the tree with HA sequences from human cases are in bold. Bootstraps greater than 50 generated from 1,000 replicates are shown at branch nodes. The scale bar represents the number of nucleotide substitutions per site.