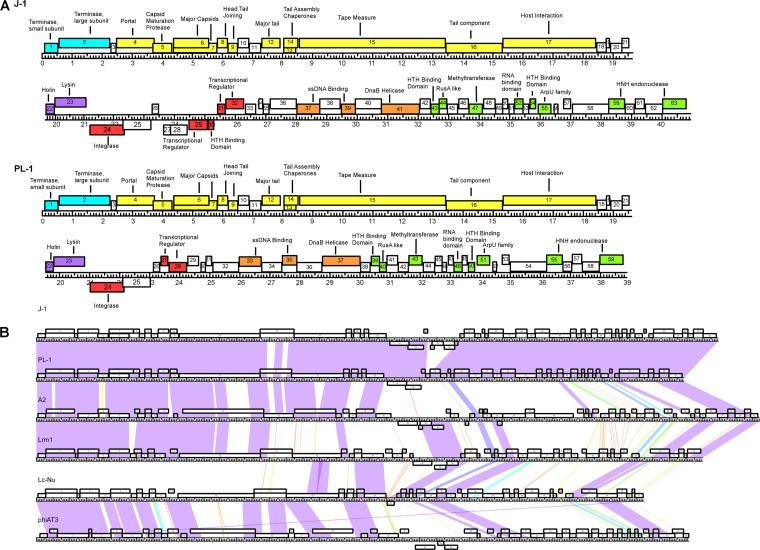
FIG 2.
(A) Annotated genome maps of bacteriophages J-1 and PL-1. The viral genomes of J-1 and PL-1 are represented in four tiers, with markers spaced at 1-kbp and 100-bp intervals. The predicted genes are shown as boxes either above or below the genome, depending on whether they are rightward or leftward transcribed, respectively. Gene numbers are shown within each box. Putative genes can be divided in the following six modules: packaging (light blue), virion structure (yellow), lysis (purple), integration and immunity (red), and replication (orange). The putative proteins found in the extreme right region are colored in green, while the ORFs lacking function are white. (B) Global comparison of Lactobacillus phages. Two pairwise nucleotide alignment of phages J-1, PL-1, and related Lactobacillus phages (Lrm1, A2, phiAT3, and Lc-Nu) using Phamerator (43). The genomes are represented by horizontal lines, with putative genes shown as boxes above (transcribed rightward) or below (transcribed leftward) each genome; the number of each gene is shown within each box.