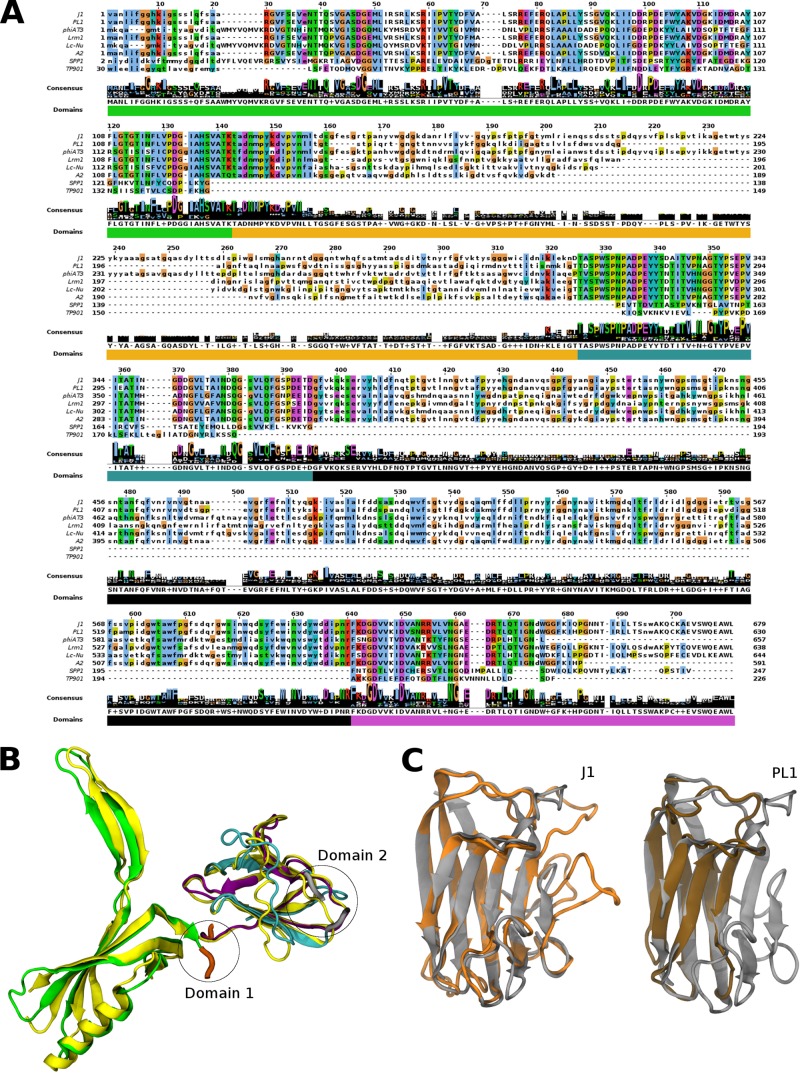
FIG 5.
gp16 alignment and structure prediction. (A) Amino acid sequence alignment of gp16 with similar proteins found in Lactobacillus phages, Orf19.1 of Spp1 and Orf46 of TP901-1. Uppercase letters correspond to aligned Pfam domain (Sypho_tail). (B) Predicted structure of gp16 of J-1 and PL-1 based on Spp1 Orf19.1 crystal structure (PDB code 2X8K, chain A). Dom1 and Dom2 correspond to the regions that could not be modeled with this PDB code. The template is shown in yellow, and the colors in the modeled structure correspond to the domains shown in panel A. (C) Predicted structure of Dom1 J-1 (orange) and Dom1 PL-1 (brown) of gp16 based on the carbohydrate-binding module (CBM) crystal structure of the endo-β-1,4-galactanase from Thermotoga maritima (PDB code 2XON, chain L) (gray).