FIG 1.
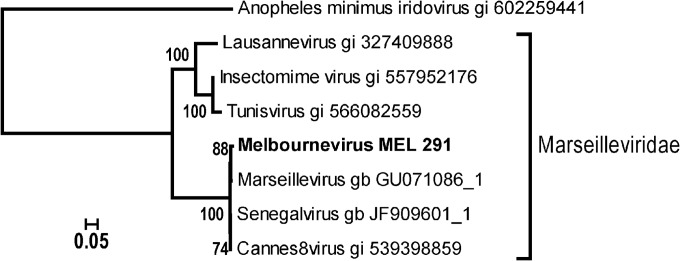
Clade structure of the Marseilleviridae. Neighbor-joining clustering was computed from 986 ungapped sites of a multiple alignment of 8 DNA polymerase catalytic-subunit amino acid sequences. The alignment and tree were computed using the Computational Biology Research Center (CBRC) server (http://www.cbrc.jp) using the Whelan and Goldman substitution model (estimated α = 1.9; 100 bootstrap resampling). The Anopheles minimus iridovirus ortholog is the best-matching homolog among viruses and is used as an outgroup. The known Marseilleviridae representatives appear to be distributed among 3 emerging subclades. The Marseillevirus subclade encompasses viruses exhibiting nearly identical genome sequences, although they were independently isolated from geographically diverse locations.
