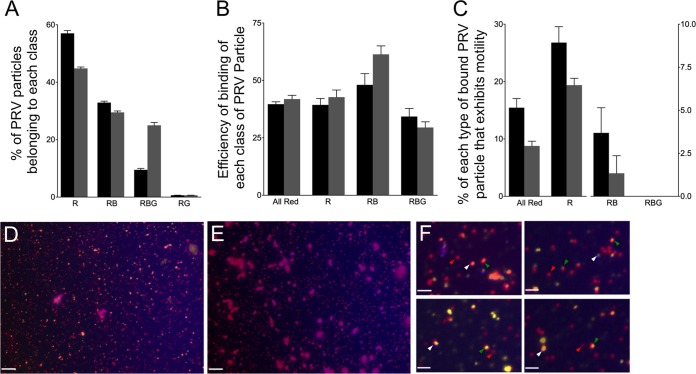
FIG 9.
Binding and motility properties of a dually fluorescently labeled PRV strain. (A) Distribution of PRV particle types in the PNS. PNS samples were prepared from cells expressing Vps4A (black bars) and Vps4A-EQ (gray bars) and examined in each fluorescence channel. (B) Efficiency of binding of each class of PRV particle to microtubules, calculated as described in the legend to Fig. 5. (C) Motility of each population of PRV particle following the addition of ATP. (A to C) Particle populations are labeled below the bars as follows: All Red, any PRV particle exhibiting at least mRFP1 fluorescence; R, PRV labeled only with mRFP1; RB, PRV particles exhibiting mRFP1 and TagBFP fluorescence; RBG, PRV particles exhibiting mRFP1, TagBFP, and GFP fluorescence. Plotted values represent means and standard deviations. Total numbers of particles examined were 2,025 (from Vps4A cells) and 1,593 (from Vps4A-EQ cells). (D) Field of PNS-derived PRV particles from a Vps4A-expressing cell stained with the lipophilic dye DiD. Image is a merge of red (capsid), blue (gD-TagBFP), and CY5 (DiD, pseudocolored yellow) channels. (E) Field similar to that shown in panel D except that DiD was omitted during staining. Scale bars in panels D and E represent 10 μm. (F) Magnification (×4) of fields similar to that shown in panel D. Indicated are red/yellow/blue particles (white arrowheads), red/yellow particles (green arrowheads), and particles that are red alone (red arrowheads). Scale bars represent 2.5 μm.