Abstract
Bacillus thuringiensis Cry3Bb, Cry3Ca, and Cry7Aa have been reported to be toxic against larvae of the genus Cylas, which are important pests of sweet potato worldwide and particularly in sub-Saharan Africa. However, relatively little is known about the processing and binding interactions of these coleopteran-specific Cry proteins. The aim of the present study was to determine whether Cry3Bb, Cry3Ca, and Cry7Aa proteins have shared binding sites in Cylas puncticollis to orient the pest resistance strategy by genetic transformation. Interestingly, processing of the 129-kDa Cry7Aa protoxin using commercial trypsin or chymotrypsin rendered two fragments of about 70 kDa and 65 kDa. N-terminal sequencing of the trypsin-activated Cry7Aa fragments revealed that processing occurs at Glu47 for the 70-kDa form or Ile88 for the 65-kDa form. Homologous binding assays showed specific binding of the two Cry3 proteins and the 65-kDa Cry7Aa fragment to brush border membrane vesicles (BBMV) from C. puncticollis larvae. The 70-kDa fragment did not bind to BBMV. Heterologous-competition assays showed that Cry3Bb, Cry3Ca, and Cry7Aa (65-kDa fragment) competed for the same binding sites. Hence, our results suggest that pest resistance mediated by the alteration of a shared Cry receptor binding site might render all three Cry toxins ineffective.
INTRODUCTION
Coleoptera comprise the largest order of insects, with large differences among families and species. Some of these species are important crop pests causing losses that are particularly damaging in developing countries. Sweet potato weevils of the genus Cylas (Coleoptera: Brentidae) are considered to be globally the most important pests of sweet potato [Ipomea batatas (L.) Lam.] (1). Cylas puncticollis (Boheman) and Cylas brunneus (Fabricius) occur in East Africa, whereas Cylas formicarius (Summers) is found in both tropical and subtropical areas worldwide (2, 3). Even small populations of sweet potato weevils can produce crop losses of up to 60 to 100% during the dry season (1). The adults lay eggs in the storage root, where larva tunneling leads to rotting, rendering it unsuitable for consumption (4). Current control strategies for these insect pests have not been effective, justifying the need for new control methods such as the use of insecticidal Cry proteins from Bacillus thuringiensis (5–7).
Cry proteins have been employed successfully worldwide to control insect pests used in formulated sprays and, more recently, expressed in transgenic crops. Since the commercialization in 1996 of the first genetically modified crop expressing a B. thuringiensis Cry toxin gene (Bt crop), the number of crops that have been transformed to express other Cry proteins has been steadily increasing (8). In addition, Bt crops are harmless to nontarget invertebrates, which are generally more abundant in Bt crop fields than when managed with chemical insecticides (9). Hence, the use of genetically engineered sweet potato plants expressing Cry toxins could control these coleopteran pests, as Bt crops have been shown to effectively control stem borers, ear feeders, and rootworms (10). The first attempt to develop a Bt sweet potato expressing Cry3A did not fully control C. formicarius due to low accumulation of the Cry protein (6, 7). Similar results were observed recently when Cry7Aa was expressed in the storage root (11). Coexpression of several Cry toxins may be an alternative to control this pest.
Molecular mechanisms that mediate the toxic activity of Cry proteins have been extensively studied, especially for Cry1A proteins active against lepidopteran larvae (12). Much less is known for coleopteran-active Cry proteins. The mode of action of B. thuringiensis Cry proteins is a multistep process that starts with the ingestion of the proteins by the larvae. Cry proteins are then solubilized in the midgut environment, and the protoxins are processed by midgut proteases into smaller fragments. The active proteins bind to specific receptors in the midgut brush border membrane of susceptible larvae and subsequently insert into the membrane, producing pores that lead to an osmotic imbalance (13, 14) and/or trigger a cell death mechanism (15, 16). In both cases, the final consequence is cell lysis and insect death (17).
The beetle-specific Cry3Bb, Cry3Ca, and Cry7Aa proteins have been described as being active for the control of C. puncticollis (5), although little is known about the interaction of these proteins in the midgut of this insect. Recently, the cry3Ca and cry7Aa genes have been used independently to transform several sweet potato varieties with the aim of controlling sweet potato weevils (11, 18). In addition, a modified Cry3Bb1 protein is expressed in transgenic corn hybrids, which are commercially available (YieldGard), to control corn rootworm larvae. The first generation of Bt crops was based on the expression of one single Cry protein. Nowadays, the trend is the combination of two or more genes expressing proteins that do not share binding sites in order to delay the evolution of resistance in target insect populations (19–21). Therefore, the aim of the present study was to assess whether Cry3Bb, Cry3Ca, and Cry7Aa proteins share binding sites in C. puncticollis. This knowledge will help in the design of insect resistance management strategies to preserve the long-term use of Bt sweet potato crops currently under development.
MATERIALS AND METHODS
Insects.
Last-instar larvae were harvested from infested sweet potato roots and immediately frozen in liquid nitrogen at the National Crops Resources Research Institute (NaCRRI), Namulonge, Uganda. These samples were then shipped on dry ice to the laboratory of the Universitat de València and stored at −80°C upon arrival.
For the collection of C. puncticollis midgut fluid, third-instar larvae were combined and centrifuged at 4,500 × g for 20 min. The supernatant was then collected, kept at −20°C until shipment, and sent frozen to the laboratory of the Universitat de València.
Bacillus thuringiensis Cry protein preparation.
Cry3Bb, Cry3Ca, or Cry7Aa proteins were obtained from B. thuringiensis strains BTS2260AA, BTS02109P, and BTS137J, respectively (provided by Bayer CropScience, Ghent, Belgium). The strains were grown in CCY medium (22) for 24 to 48 h at 29°C. After complete sporulation, spores and crystals were harvested and washed three times with 1 M NaCl–10 mM EDTA and twice with 10 mM KCl. Cry3 proteins were solubilized and activated by bovine pancreas α-chymotrypsin (type I-S) (Sigma-Aldrich), as described previously by Rausell et al. (23). Cry7Aa crystals were solubilized in 50 mM carbonate buffer (Na2CO3-NaHCO3, pH 10.5)–10 mM dithiothreitol (DTT) for 2 h at room temperature. The Cry7Aa protoxin was activated with either bovine pancreas trypsin (type I) (Sigma-Aldrich) (1:5 [wt/wt] protease/protoxin ratio) or bovine pancreas α-chymotrypsin (2:1 [wt/wt] protease/protoxin ratio) for 3 h at 37°C. Alternatively, Cry7Aa protoxin was processed with midgut fluid from C. puncticollis. Prior to use, the midgut fluid was thawed on ice and centrifuged at 13,000 × g for 10 min at 4°C, and the supernatant containing soluble proteases was recovered. The protein concentration of the gut fluid was determined by the method of Bradford (24), using bovine serum albumin (BSA) as a standard. The Cry7Aa protein was incubated with gut juice (1:3 [wt/wt] total protein gut juice/protoxin ratio) for 16 h at 30°C. Dilutions of the gut fluid were done in phosphate-buffered saline (PBS) (10 mM Na2HPO4, 1 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl [pH 7.4]).
At the end of the activation process, the insoluble material was removed by centrifugation (12,000 × g for 10 min), and the purity of the activated toxins was checked by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% SDS-PAGE). The activated toxins were kept at −20°C until use.
The trypsin-activated Cry7Aa sample was dialyzed in 20 mM Tris-HCl (pH 8.6) and further purified by anion-exchange chromatography in a MonoQ 5/5 column using an Äkta 100 explorer system (GE Healthcare, United Kingdom).
Brush border membrane vesicle preparation.
Brush border membrane vesicles (BBMV) from C. puncticollis whole last-instar larvae were prepared by the differential magnesium precipitation method (25), as modified by Escriche et al. (26). The protein concentration in the BBMV preparation was determined by the method of Bradford, using BSA as a standard.
Leucine aminopeptidase was used as a membrane enzymatic marker for the BBMV preparations. Leucine aminopeptidase activity was measured as described previously by Hernández et al. (27). The specific activity of the leucine aminopeptidase in the BBMV preparation (1.5 ± 0.1 μmol min−1 mg−1 protein [mean ± standard deviation {SD}]) was enriched approximately 10-fold relative to the crude homogenate (0.15 ± 0.01 μmol min−1 mg−1 protein [mean ± SD]).
Labeling of Cry proteins.
Activated Cry proteins were biotinylated by using a protein biotinylation kit (GE Healthcare) according to the manufacturer's instructions. Prior to labeling, proteins were dialyzed overnight at 4°C in 40 mM Na2CO3-NaHCO3 buffer (pH 8.6). After biotin labeling, the mixture was loaded onto a PD10 desalting column (GE Healthcare) equilibrated with PBS. The eluted fractions were analyzed by 12% SDS-PAGE.
Binding assays.
Prior to use, BBMV were centrifuged for 10 min at 16,000 × g and suspended in binding buffer (PBS, 0.1% BSA [pH 7.4]). Competition experiments were performed by incubating 50 ng of biotinylated Cry3 proteins with 5 μg of BBMV or by incubating 100 ng of biotinylated Cry7Aa protein with 20 μg of BBMV in binding buffer. Incubations were carried out for 1 h at 25°C in the absence or presence of an excess of unlabeled proteins (50- to 200-fold excess) in a final volume of 100 μl. After incubation, samples were centrifuged at 16,000 × g for 10 min, and the pellets were washed with 500 μl of ice-cold binding buffer. The final pellets, containing the bound biotinylated proteins, were suspended in 10 μl of the same buffer and analyzed by 12% SDS-PAGE. The separated proteins were electroblotted onto a nitrocellulose membrane (Hybond-ECL; GE Healthcare). Biotinylated proteins were visualized after probing with streptavidin-conjugated horseradish peroxidase (1:2,000 dilution) with a chemiluminescence detection procedure (RPN2109; GE Healthcare), using an ImageQuant LAS400 image analyzer. Each competition experiment was repeated a minimum of three times.
N-terminal sequence analysis of the Cry7Aa trypsin fragments.
The chromatography-purified trypsin-activated Cry7Aa fragments were subjected to 10% SDS-PAGE and electroblotted onto a polyvinylidene difluoride (PVDF) membrane. Protein bands were then cut out and sent for sequencing. N-terminal amino acid sequencing was performed by using a Procise 492 cLC (Applied Biosystems) at Alphalyse A/S (Odense, Denmark).
RESULTS
Proteolytic processing of Cry protoxins.
Chymotrypsin treatment of the Cry3Bb and Cry3Ca protoxins (approximately 73 kDa each) produced polypeptides with masses of about 55 kDa and 53 kDa, respectively (data not shown). On the other hand, processing of the Cry7Aa protoxin (129 kDa) with trypsin and chymotrypsin provided similar proteolytic patterns, with two main fragments of about 70 and 65 kDa (Fig. 1A). In contrast, the activation of the Cry7Aa protoxin by gut fluid from C. puncticollis showed a single band with a mass of about 65 kDa (Fig. 1B).
FIG 1.
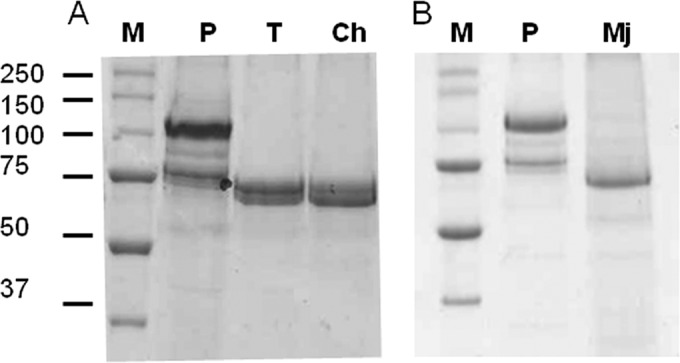
SDS-PAGE (12%) analysis of the protease-resistant fragments obtained after different proteolytic processing of the solubilized Cry7Aa protoxin. (A) Activation with commercial trypsin (T) or chymotrypsin (Ch). (B) Activation with midgut juice from C. puncticollis (Mj). M, molecular mass marker (in kDa); P, untreated solubilized Cry7Aa protein.
Binding of Cry3Bb, Cry3Ca, and Cry7Aa to BBMV.
Biotin-labeled Cry3Bb and Cry3Ca chymotrypsin-activated proteins bound to BBMV from C. puncticollis. Homologous competition with an excess of their respective unlabeled Cry3Bb or Cry3Ca proteins substantially reduced binding of the biotinylated Cry3 proteins, indicating that most of the binding was specific (Fig. 2A and B).
FIG 2.
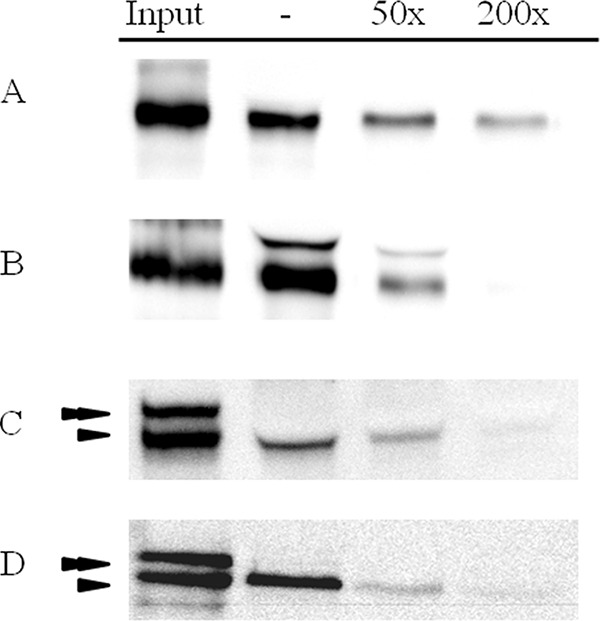
Homologous competition binding assays with BBMV from C. puncticollis. Biotinylated chymotrypsin-activated Cry3Bb (A), biotinylated chymotrypsin-activated Cry3Ca (B), biotinylated trypsin-activated Cry7Aa (C), or biotinylated chymotrypsin-activated Cry7Aa (D) was incubated with (50× to 200×) or without (−) an excess of unlabeled activated protein. Input indicates biotinylated Cry proteins. The double and single arrowheads indicate the 70-kDa and 65-kDa fragments, respectively, obtained after trypsin or chymotrypsin processing of the Cry7Aa protein.
The mixture of 70- and 65-kDa bands, resulting after either trypsin or chymotrypsin treatment of Cry7Aa, was labeled with biotin. Homologous-competition assays showed that the 65-kDa fragment was able to bind and that this interaction was specific, since it was competed with a 200-fold excess of unlabeled protein (Fig. 2C and D). In contrast, no binding was observed for the 70-kDa fragment. To determine whether the lack of binding of the 70-kDa fragment is due to its in vitro processing to the 65-kDa fragment during incubation with BBMV, both proteins were separated, individually labeled, and tested for binding. The results showed that only the purified 65-kDa fragment bound specifically to BBMV (Fig. 3C and D), whereas the 70-kDa fragment did not bind to BBMV (Fig. 4C).
FIG 3.
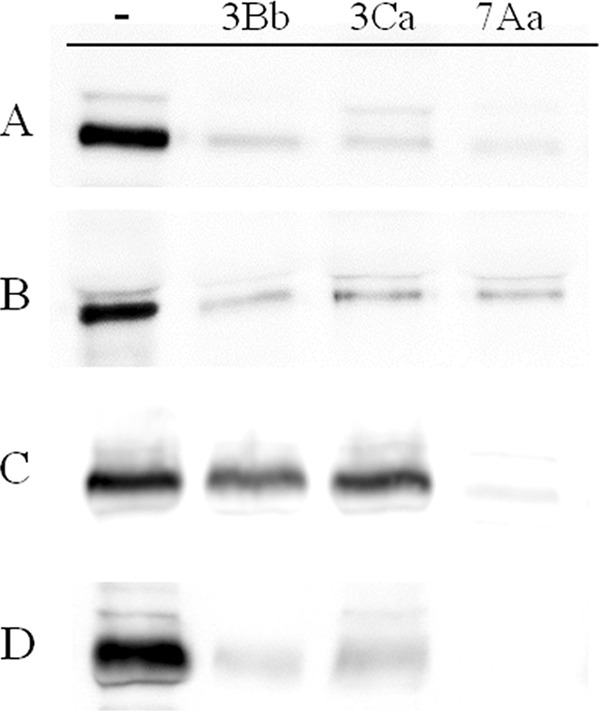
Binding of biotinylated Cry3Bb (A), Cry3Ca (B), and Cry7Aa (65-kDa fragment) (C and D) proteins to C. puncticollis BBMV in the absence of competitor (−) or in the presence of a 100-fold (A to C) or a 200-fold (D) excess of competitor (lanes labeled 3Bb, 3Ca, and 7Aa [65-kDa fragment]).
FIG 4.
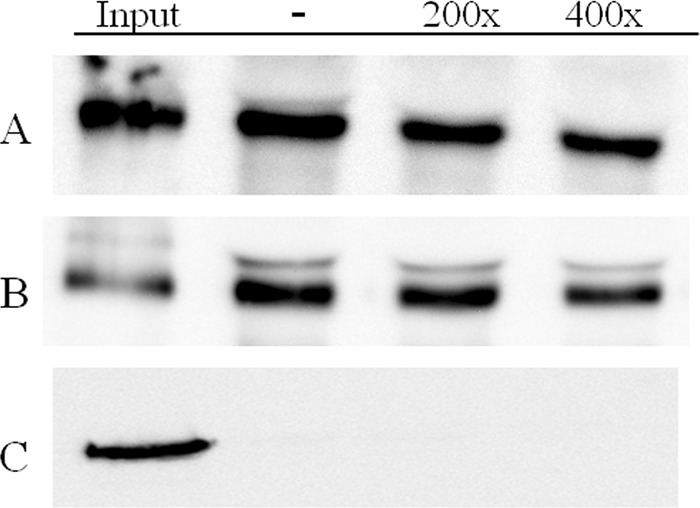
Binding of biotinylated Cry3Bb (A), Cry3Ca (B), and Cry7Aa (70-kDa fragment) (C) proteins to C. puncticollis BBMV in the absence of competitor (−) or in the presence of a 200- or 400-fold excess of Cry7Aa (70-kDa fragment).
Heterologous competition among Cry3Bb, Cry3Ca, and Cry7Aa.
Heterologous competition of Cry3Bb with Cry3Ca showed that both proteins were able to completely compete with the labeled protein, suggesting that both proteins share the same binding sites (Fig. 3A and B). Similarly, heterologous competition of either labeled Cry3Bb or Cry3Ca with the purified 65-kDa Cry7Aa protein showed that the latter competed with both labeled proteins (Fig. 3A and B). In all cases, the heterologous competitor was as effective as the homologous one, suggesting that there are no unshared sites for these two Cry3 proteins.
Data for heterologous binding assays with the labeled 65-kDa Cry7Aa protein and the other two Cry3 proteins are shown in Fig. 3C and D. The results showed that a 100-fold excess of unlabeled Cry7Aa (65-kDa fragment) competes with most of the bound labeled Cry7Aa, whereas only part of the labeled Cry7Aa is displaced by a 100-fold excess of either Cry3Bb or Cry3Ca. However, a 200-fold excess of both Cry3 proteins substantially reduced labeled Cry7Aa binding (Fig. 3D). These results suggest that Cry3Bb and Cry3Ca recognize the same binding sites as Cry7Aa although with a lower affinity.
To test whether the lack of binding of the labeled 70-kDa Cry7Aa protein is due to alteration of the binding epitope during the labeling process, the unlabeled fragment was used as a heterologous competitor with labeled Cry3 proteins. The results showed that the purified 70-kDa Cry7Aa protein was unable to compete for the Cry3Bb or Cry3Ca binding sites, even when a 400-fold excess was used (Fig. 4).
N-terminal sequence analysis of Cry7Aa trypsin fragments.
The N-terminal sequence of the 70-kDa fragment obtained after trypsin treatment was EQPEA, corresponding to Glu47, whereas the N-terminal sequence of the 65-kDa trypsin-activated fragment was IAGLL (Ile88).
In silico analysis of the Cry7Aa protein sequence compared to other well-studied three-domain Cry proteins suggested that the 70-kDa fragment is produced by cleavage before the beginning of helix α1, whereas the 65-kDa fragment comes from cleavage at the end of helix α2.
DISCUSSION
Despite the large number of B. thuringiensis Cry proteins that have been described (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/), only a few are known to be active against Cylas spp. (5–7). Since relatively little is known about the processing and binding interactions of beetle-specific Cry proteins, we studied the interactions of three active Cry proteins (Cry3Bb, Cry3Ca, and Cry7Aa) against the sweet potato weevil C. puncticollis. This knowledge will also guide future attempts to produce Bt sweet potato with multiple cry genes.
It is well known that Cry protoxins are processed to active proteins by midgut proteases to exert their insecticidal activity (12). We have shown that activation of solubilized Cry3Bb and Cry3Ca with chymotrypsin renders the expected polypeptides with masses of about 55 kDa and 53 kDa, respectively (23, 28). Processing of the Cry7Aa protoxin with either trypsin or chymotrypsin provided two fragments of about 70 kDa and 65 kDa, whereas activation by C. puncticollis gut fluid generated a single 65-kDa fragment. Previously reported results for the proteolytic processing of Cry7Aa slightly differed from the results obtained in this study. A single 72-kDa fragment was reported after activation with either Colorado potato beetle gut juice or trypsin (29), whereas Peferoen et al. (30) reported a single 66-kDa fragment after trypsin treatment. These differences could be attributed to differences in the experimental conditions used. The occurrence of two fragments after in vitro proteolytic processing was also described for another member of the Cry7 family (Cry7Ab3) and for members of other families, such as Cry1Ab, Cry3Aa, Cry8D, and Cry9Ca (31–37).
The present work is the first report of binding assays of beetle-active Cry proteins with C. puncticollis BBMV. Although previous studies have shown the ability of some Cry3 proteins to bind to BBMV from Coleoptera (23, 38–41), this is the first time that Cry7 has been included in binding studies. Homologous-competition assays carried out with chymotrypsin-activated Cry3Bb and Cry3Ca proteins demonstrated that the binding of these two proteins to C. puncticollis BBMV is specific. Furthermore, homologous-competition experiments with either trypsin- or chymotrypsin-activated Cry7Aa protein, consisting of a mixture of the 70- and 65-kDa fragments, showed that only the 65-kDa fragment was able to bind to BBMV from C. puncticollis and that this binding was specific.
Heterologous-competition binding experiments have provided models for binding sites to predict or to explain patterns of cross-resistance (42–45). The results with C. puncticollis could be interpreted as that the three proteins bind to a single binding site, although Cry3Bb and Cry3Ca bind with lower affinity. Alternatively, the three proteins share a binding site with similar affinity, and Cry7Aa has an additional binding site to which Cry3 proteins bind with lower affinity. In either case, an alteration of the shared binding site in C. puncticollis might confer resistance to these three proteins. The occurrence of shared binding sites for Cry3 proteins (Cry3Aa, Cry3Ba, and Cry3Ca) was previously described in a study of BBMV isolated from Colorado potato beetle (23). Nevertheless, this is the first study that demonstrated the occurrence of shared binding sites between Cry3 and Cry7Aa proteins.
Cry7Aa belongs to the 130-kDa Cry protein family (29). In general, processing of 130-kDa Cry proteins involves the removal of 25 to 60 residues from the N-terminal end and approximately half of the remaining protein from the C terminus (46, 47). Sequencing of the N-terminal end of the purified trypsin-activated Cry7Aa fragments revealed that the 70-kDa fragment starts at Glu47, whereas the 65-kDa fragment is produced after the removal of the N-terminal sequence up to Ile88. The cleavage of the N terminus has been described as a critical step for some Cry proteins to be considered active proteins (48–53). Indirect evidence based on the structure of the unprocessed Cry2Aa protein revealed that the N-terminal region masks a region of the protein that could be involved in the interaction between the protein and the membrane of the target insect (54). Based on these results, the differences in the binding abilities observed for the 70-kDa and 65-kDa Cry7Aa fragments could be due to the differences in the N-terminal sequences. Similar results were described for the beetle-specific Cry8Da protein, where processing by Japanese beetle gut juice rendered two different polypeptides and only the small fragment bound to BBMV proteins (37).
As the 70-kDa trypsin-activated fragment is unable to bind to C. puncticollis BBMV, it was interesting to observe that it was found to be active against C. puncticollis larvae (Runyararo J. Rukarwa, personal communication). These results, along with the fact that the Cry7Aa protoxin is processed to the 65-kDa fragment with C. puncticollis gut fluid, suggest that the 70-kDa fragment is further processed in vivo to the 65-kDa fragment. These results are highly relevant to the Bt sweet potato currently under development, because the transgene expresses the toxic fragment starting at position 59 upstream of the proteolytic cleavage site yielding the 65-kDa fragment (18). Thus, knowledge about the processing of the Cry proteins (at the N and C termini) may assist in the design of constructs for plant expression.
In summary, the current study provides evidence of the importance of proper proteolytic activation for the in vitro ability of Cry7Aa to bind to the midgut receptors of C. puncticollis. Furthermore, based on the results of binding-site interactions, the development of cross-resistance among Cry3Bb, Cry3Ca, and Cry7Aa proteins due to binding-site modification appears to be possible for C. puncticollis. Thus, from a resistance management standpoint, combinations of Cry3Bb, Cry3Ca, or Cry7Aa proteins do not seem to be suitable for the development of Bt sweet potato plants.
ACKNOWLEDGMENTS
We thank Jeroen Van Rie (Bayer CropScience NV) for providing the B. thuringiensis strains that produced the Cry3Bb, Cry3Ca, and Cry7Aa proteins and for his helpful discussions, Runyararo J. Rukarwa for providing the C. puncticollis larvae and the midgut fluid from this insect species, and the National Crop Resources Research Institute in Uganda.
This work was supported by the Bill and Melinda Gates Foundation through the Sweetpotato Action for Security and Health in Africa (SASHA) project, with additional support for the weevil activities in Uganda from the U.S. Agency for International Development (USAID).
Footnotes
Published ahead of print 26 September 2014
REFERENCES
- 1.Chalfant RB, Jansson RK, Seal DR, Schalk JM. 1990. Ecology and management of sweet potato insects. Annu. Rev. Entomol. 35:157–180. 10.1146/annurev.en.35.010190.001105. [DOI] [Google Scholar]
- 2.Smit NEJM, van Huis A. 1998. Biology of the African sweetpotato weevil species Cylas puncticollis (Boheman) and C. brunneus (Fabricius) (Coleoptera: Apionidae). Int. J. Trop. Insect Sci. 18:93–100. 10.1017/S1742758400007712. [DOI] [Google Scholar]
- 3.Smith TP, Hammond AM. 2006. Comparative susceptibility of sweetpotato weevil (Coleoptera: Brentidae) to selected insecticides. J. Econ. Entomol. 99:2024–2029. 10.1603/0022-0493-99.6.2024. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen KA. 2009. Sweetpotato insects: identification, biology and management, p 157–184 In Loebensteinand G, Thottappilly G. (ed), The sweetpotato. Springer-Verlag Inc, New York, NY. [Google Scholar]
- 5.Ekobu M, Solera M, Kyamanywa S, Mwanga RO, Odongo B, Ghislain M, Moar WJ. 2010. Toxicity of seven Bacillus thuringiensis Cry proteins against Cylas puncticollis and Cylas brunneus (Coleoptera: Brentidae) using a novel artificial diet. J. Econ. Entomol. 103:1493–1502. 10.1603/EC09432. [DOI] [PubMed] [Google Scholar]
- 6.García R, Morán R, Mena J, Somontes D, Pimentel E, Zaldúa Z, López A, García M. 1999. Sweetpotato (Ipomea batatas L.) regeneration and transformation technology to provide weevil (Cylas formicarius) resistance. Field trial results, p 112–117 In Arencibia AD. (ed), Plant genetic engineering: towards the third millennium. Elsevier Science BV, Amsterdam, Netherlands. [Google Scholar]
- 7.Moran R, García R, López A, Zaldua Z, Mena J, García M, Armas R, Somonte D, Rodríguez J, Gómez M, Pimentel E. 1998. Transgenic sweet potato plants carrying the delta-endotoxin gene from Bacillus thuringiensis var. tenebrionis. Plant Sci. 139:175–184. 10.1016/S0168-9452(98)00179-4. [DOI] [Google Scholar]
- 8.James C. 2013. Global status of commercialized biotech/GM crops: 2013. ISAAA brief 46 ISAAA, Ithaca, NY. [Google Scholar]
- 9.Marvier M, McCreedy C, Regetz J, Kareiva P. 2007. A meta-analysis of effects of Bt cotton and maize on nontarget invertebrates. Science 316:1475–1477. 10.1126/science.1139208. [DOI] [PubMed] [Google Scholar]
- 10.Shelton AM, Zhao JZ, Roush RT. 2002. Economic, ecological, food safety, and social consequences of the deployment of Bt transgenic plants. Annu. Rev. Entomol. 47:845–881. 10.1146/annurev.ento.47.091201.145309. [DOI] [PubMed] [Google Scholar]
- 11.Rukarwa RJ, Prentice K, Ormachea M, Kreuze JF, Tovar J, Mukasa SB, Ssemakula G, Mwanga ROM, Ghislain M. 2013. Evaluation of bioassays for testing Bt sweetpotato events against sweetpotato weevils. African Crop Sci. J. 21:235–244. [Google Scholar]
- 12.Pardo-López L, Soberón M, Bravo A. 2013. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 37:3–22. 10.1111/j.1574-6976.2012.00341.x. [DOI] [PubMed] [Google Scholar]
- 13.Knowles BH, Ellar DJ. 1987. Colloid-osmotic lysis is a general feature of the mechanism of action of Bacillus thuringiensis δ-endotoxins with different insect specificity. Biochim. Biophys. Acta 924:509–518. 10.1016/0304-4165(87)90167-X. [DOI] [Google Scholar]
- 14.Bravo A, Gómez I, Conde J, Muñoz-Garay C, Sánchez J, Miranda R, Zhuang M, Gill SS, Soberón M. 2004. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim. Biophys. Acta 1667:38–46. 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Candas M, Griko NB, Rose-Young L, Bulla LA. 2005. Cytotoxicity of Bacillus thuringiensis Cry1Ab toxin depends on specific binding of the toxin to the cadherin receptor BT-R1 expressed in insect cells. Cell Death Differ. 12:1407–1416. 10.1038/sj.cdd.4401675. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Candas M, Griko NB, Taussig R, Bulla LA. 2006. A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc. Natl. Acad. Sci. U. S. A. 103:9897–9902. 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vachon V, Laprade R, Schwartz JL. 2012. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: a critical review. J. Invertebr. Pathol. 111:1–12. 10.1016/j.jip.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Kreuze JF, Valkonen JPT, Ghislain M. 2009. Genetic engineering, p 41–63 In Loebebstein G, Thottappilly G. (ed), The sweetpotato. Springer-Verlag Inc, New York, NY. [Google Scholar]
- 19.Ferré J, Van Rie J, MacIntosh SC. 2008. Insecticidal genetically modified crops and insect resistance management (IRM), p 41–85 In Romeis J, Shelton AM, Kennedy GG. (ed), Integration of insect-resistant genetically modified crops within IPM programs. Springer, Dordrecht, Netherlands. [Google Scholar]
- 20.Roush RT. 1998. Two-toxin strategies for management of insecticidal transgenic crops: can pyramiding succeed where pesticide mixtures have not? Philos. Trans. R. Soc. Lond. B Biol. Sci. 353:1777–1786. 10.1098/rstb.1998.0330. [DOI] [Google Scholar]
- 21.Zhao JZ, Cao J, Li Y, Collins HL, Roush RT, Earle ED, Shelton AM. 2003. Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat. Biotechnol. 21:1493–1497. 10.1038/nbt907. [DOI] [PubMed] [Google Scholar]
- 22.Stewart GSAB, Johnstone K, Hagelberg E, Ellar DJ. 1981. Commitment of bacterial spores to germinate. Biochem. J. 198:101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rausell C, García-Robles I, Sánchez J, Muñoz-Garay C, Martínez-Ramírez AC, Real MD, Bravo A. 2004. Role of toxin activation on binding and pore formation activity of the Bacillus thuringiensis Cry3 toxins in membranes of Leptinotarsa decemlineata (Say). Biochim. Biophys. Acta 1660:99–105. 10.1016/j.bbamem.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Wolfersberger MG, Luthy P, Parenti P, Parenti P, Sacchi VF, Giordana B, Hanozet GM. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. A Physiol. 86A:301–308. [Google Scholar]
- 26.Escriche B, Silva FS, Ferré J. 1995. Testing suitability of brush border membrane vesicles prepared from whole larvae from small insects for binding studies with Bacillus thuringiensis CrylA(b) crystal protein. J. Invertebr. Pathol. 65:318–320. 10.1006/jipa.1995.1051. [DOI] [Google Scholar]
- 27.Hernández CS, Rodrigo A, Ferré J. 2004. Lyophilization of lepidopteran midguts: a preserving method for Bacillus thuringiensis toxin binding studies. J. Invertebr. Pathol. 85:182–187. 10.1016/j.jip.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Park Y, Abdullah MA, Taylor MD, Rahman K, Adang MJ. 2009. Enhancement of Bacillus thuringiensis Cry3Aa and Cry3Bb toxicities to coleopteran larvae by a toxin-binding fragment of an insect cadherin. Appl. Environ. Microbiol. 75:3086–3092. 10.1128/AEM.00268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert B, Höfte H, Annys K, Jansens S, Soetaert P, Peferoen M. 1992. Novel Bacillus thuringiensis insecticidal crystal protein with a silent activity against coleopteran larvae. Appl. Environ. Microbiol. 58:2536–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peferoen M, Lambert B, Joos H. January 1994. Plants transformed to produce Bacillus thuringiensis insecticidal proteins. European patent 0 458 819 B1. [Google Scholar]
- 31.Carroll J, Convents D, Van Damme J, Boets A, Van Rie J, Ellar DJ. 1997. Intramolecular proteolytic cleavage of Bacillus thuringiensis Cry3A delta-endotoxin may facilitate its coleopteran toxicity. J. Invertebr. Pathol. 70:41–49. 10.1006/jipa.1997.4656. [DOI] [PubMed] [Google Scholar]
- 32.Lambert B, Buysse L, Decock C, Jansens S, Piens C, Saey B, Seurinck J, Van Audenhove K, Van Rie J, Van Vliet A, Peferoen M. 1996. A Bacillus thuringiensis insecticidal crystal protein with a high activity against members of the family Noctuidae. Appl. Environ. Microbiol. 62:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranda R, Zamudio FZ, Bravo A. 2001. Processing of Cry1Ab delta-endotoxin from Bacillus thuringiensis by Manduca sexta and Spodoptera frugiperda midgut proteases: role in protoxin activation and toxin inactivation. Insect Biochem. Mol. Biol. 31:1155–1163. 10.1016/S0965-1748(01)00061-3. [DOI] [PubMed] [Google Scholar]
- 34.Song P, Wang Q, Nangong Z, Su J, Ge D. 2012. Identification of Henosepilachna vigintioctomaculata (Coleoptera: Coccinellidae) midgut putative receptor for Bacillus thuringiensis insecticidal Cry7Ab3 toxin. J. Invertebr. Pathol. 109:318–322. 10.1016/j.jip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Walters FS, Stacy CM, Lee MK, Palekar N, Chen JS. 2008. An engineered chymotrypsin/cathepsin G site in domain I renders Bacillus thuringiensis Cry3A active against Western corn rootworm larvae. Appl. Environ. Microbiol. 74:367–374. 10.1128/AEM.02165-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi T, Sahara K, Bando H, Asano S. 2008. Discovery of a novel Bacillus thuringiensis Cry8D protein and the unique toxicity of the Cry8D-class proteins against scarab beetles. J. Invertebr. Pathol. 99:257–262. 10.1016/j.jip.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi T, Sahara K, Bando H, Asano S. 2010. Intramolecular proteolytic nicking and binding of Bacillus thuringiensis Cry8Da toxin in BBMVs of Japanese beetles. J. Invertebr. Pathol. 105:243–247. 10.1016/j.jip.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Belfiore CJ, Vadlamudi RK, Osman YA, Bulla LA., Jr 1994. A specific binding protein from Tenebrio molitor for the insecticidal toxin of Bacillus thuringiensis subsp. tenebrionis. Biochem. Biophys. Res. Commun. 200:359–364. 10.1006/bbrc.1994.1456. [DOI] [PubMed] [Google Scholar]
- 39.Carroll J, Convents D, Van Damme J, Boets A, Van Rie J, Ellar DJ. 1997. Intramolecular proteolytic cleavage of Bacillus thuringiensis Cry3A delta-endotoxin may facilitate its coleopteran toxicity. J. Invertebr. Pathol. 70:41–49. 10.1006/jipa.1997.4656. [DOI] [PubMed] [Google Scholar]
- 40.Martínez-Ramírez AC, Real MD. 1996. Proteolytic processing of Bacillus thuringiensis CryIIIA toxin and specific binding to brush-border membrane vesicles of Leptinotarsa decemlineata (Colorado potato beetle). Pestic. Biochem. Physiol. 54:115–122. 10.1006/pest.1996.0015. [DOI] [Google Scholar]
- 41.Li H, Olson M, Lin G, Hey T, Tan SY, Narva KE. 2013. Bacillus thuringiensis Cry34Ab1/Cry35Ab1 interactions with Western corn rootworm midgut membrane binding sites. PLoS One 8:e53079. 10.1371/journal.pone.0053079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballester V, Granero F, Tabashnik BE, Malvar T, Ferré J. 1999. Integrative model for binding of Bacillus thuringiensis toxins in susceptible and resistant larvae of the diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 65:1413–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.González-Cabrera J, Escriche B, Tabashnik BE, Ferré J. 2003. Binding of Bacillus thuringiensis toxins in resistant and susceptible strains of pink bollworm (Pectinophora gossypiella). Insect Biochem. Mol. Biol. 33:929–935. 10.1016/S0965-1748(03)00099-7. [DOI] [PubMed] [Google Scholar]
- 44.Hernández CS, Ferré J. 2005. Common receptor for Bacillus thuringiensis toxins Cry1Ac, Cry1Fa, and Cry1Ja in Helicoverpa armigera, Helicoverpa zea, and Spodoptera exigua. Appl. Environ. Microbiol. 71:5627–5629. 10.1128/AEM.71.9.5627-5629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernández-Rodríguez CS, Hernández-Martínez P, Van Rie J, Escriche B, Ferré J. 2013. Shared midgut binding sites for Cry1A.105, Cry1Aa, Cry1Ab, Cry1Ac and Cry1Fa proteins from Bacillus thuringiensis in two important corn pests, Ostrinia nubilalis and Spodoptera frugiperda. PLoS One 8:e68164. 10.1371/journal.pone.0068164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Maagd RA, Bravo A, Berry C, Crickmore N, Schnepf HE. 2003. Structure, diversity and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu. Rev. Genet. 37:409–433. 10.1146/annurev.genet.37.110801.143042. [DOI] [PubMed] [Google Scholar]
- 47.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bravo A, Sánchez J, Kouskoura T, Crickmore N. 2002. N-terminal activation is an essential early step in the mechanism of action of the Bacillus thuringiensis Cry1Ac insecticidal toxin. J. Biol. Chem. 277:23985–23987. 10.1074/jbc.C200263200. [DOI] [PubMed] [Google Scholar]
- 49.Franklin MT, Nieman CL, Janmaat AF, Soberón M, Bravo A, Tabashnik BE, Myers JH. 2009. Modified Bacillus thuringiensis toxins and a hybrid B. thuringiensis strain counter greenhouse-selected resistance in Trichoplusia ni. Appl. Environ. Microbiol. 75:5739–5741. 10.1128/AEM.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gómez I, Oltean DI, Gill SS, Bravo A, Soberón M. 2001. Mapping the epitope in cadherin-like receptors involved in Bacillus thuringiensis Cry1A toxin interaction using phage display. J. Biol. Chem. 276:28906–28912. 10.1074/jbc.M103007200. [DOI] [PubMed] [Google Scholar]
- 51.Gómez I, Sánchez J, Miranda R, Bravo A, Soberón M. 2002. Cadherin-like receptor binding facilitates proteolytic cleavage of helix alpha-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 513:242–246. 10.1016/S0014-5793(02)02321-9. [DOI] [PubMed] [Google Scholar]
- 52.Lightwood DJ, Ellar DJ, Jarrett P. 2000. Role of proteolysis in determining potency of Bacillus thuringiensis Cry1Ac δ-endotoxin. Appl. Environ. Microbiol. 66:5174–5181. 10.1128/AEM.66.12.5174-5181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajagopal R, Arora N, Sivakumar S, Rao NG, Nimbalkar SA, Bhatnagar RK. 2009. Resistance of Helicoverpa armigera to Cry1Ac toxin from Bacillus thuringiensis is due to improper processing of the protoxin. Biochem. J. 419:309–316. 10.1042/BJ20081152. [DOI] [PubMed] [Google Scholar]
- 54.Morse RJ, Yamamoto T, Stroud RM. 2001. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure 9:409–417. 10.1016/S0969-2126(01)00601-3. [DOI] [PubMed] [Google Scholar]


