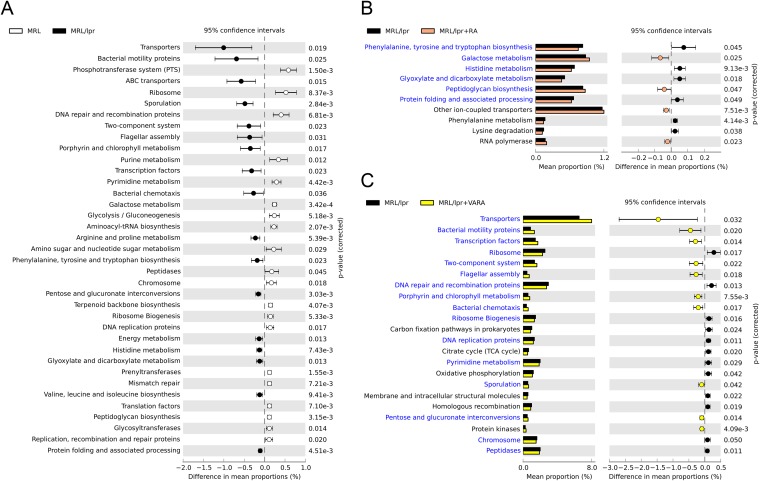
FIG 4.
Bacterial metagenomes predicted from microbial community identities. Gene functional categories were from level 3 of KEGG pathways. Gene functions with a significant difference are shown (P < 0.05). (A) Differences between MRL control and lupus-prone MRL/lpr mice. The effect size was 0.10%. ABC transporters, ATP-binding cassette transport systems. (B) Effect of RA treatment on lupus-prone bacterial metagenomes. The effect size was 0.02%. (C) Effect of retinol treatment (VARA) on lupus-prone bacterial metagenomes. The effect size was 0.08%.