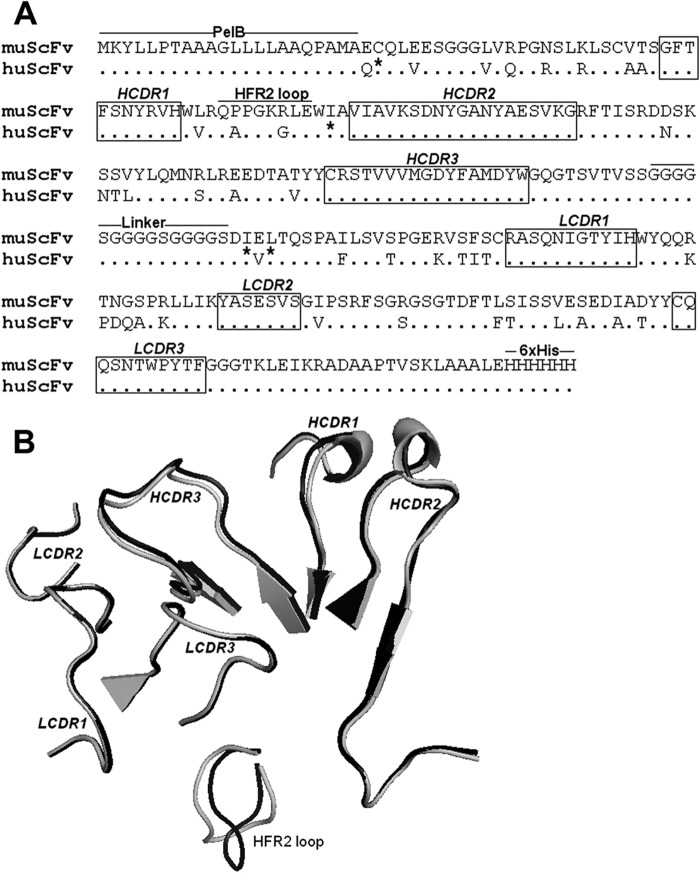
FIG 2.
Comparison of the CDR amino acid sequence and structure of muscFv1E4 with huscFv1E4. (A) Alignment of deduced amino acid sequences of muscFv1E4 and huscFv1E4. Nonidentical residues are shown. Dots indicate identical residues. The definitions of CDR residues according to the Kabat database are indicated in boxes. H, heavy chain; L, light chain; CDR, complementarity-determining region; FR, framework region. The retained murine amino acids (back mutation) are indicated by asterisks. PelB leader sequence, HFR2 loop, linker, and 6×His tag are underlined. (B) Structure alignment models of muscFv1E4 (black) and huscFv1E4 (gray). The cartoon graph displays the loop structure of CDR domains and HFR2.