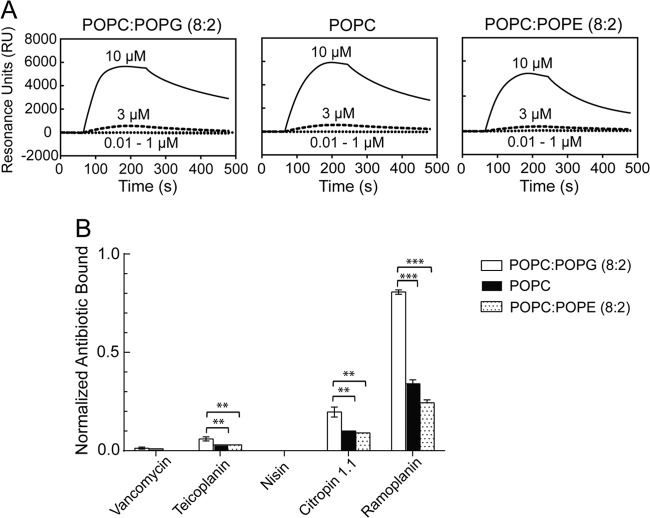
FIG 3.
(A) Typical SPR sensorgrams, showing the changes in resonance units (RU) against time upon binding of ramoplanin (0.01 to 10 μM) to lipid bilayers comprised of different lipid mixtures reconstituted on an L1 lipid capture sensor chip. Ramoplanin was injected over the lipid surface for 180 s, and the ramoplanin-lipid complex was then allowed to dissociate for 300 s. The baseline was set to zero for ease of visualization and represents the value RULipid. (B) Comparison of the binding affinities of antibiotics toward three different lipid bilayers at 10 μM, after normalization of the amount of antibiotic bound (RUBound) in SPR sensorgrams against the corresponding antibiotic molecular weight and the amount of lipid loaded on each channel of the sensor chip (RULipid), as described in Materials and Methods. Statistical comparisons of normalized antibiotic bound to the anionic (POPC:POPG, 8:2) and zwitterionic (POPC and POPC:POPE, 8:2) membranes was performed by using the two-tailed Student t test. **, P < 0.01; ***, P < 0.001. Data are means ± standard deviations (n = 3).