Abstract
The human cytomegalovirus (HCMV) immediate-early 2 (IE2) protein is a multifunctional factor essential for viral replication. IE2 modulates both viral and host gene expression, deregulates cell cycle progression, acts as an immunomodulator, and antagonizes cellular antiviral responses. Based on these facts, IE2 has been proposed as an important target for the development of innovative antiviral approaches. We previously identified the 6-aminoquinolone WC5 as a promising inhibitor of HCMV replication, and here, we report the dissection of its mechanism of action against the viral IE2 protein. Using glutathione S-transferase (GST) pulldown assays, mutagenesis, cell-based assays, and electrophoretic mobility shift assays, we demonstrated that WC5 does not interfere with IE2 dimerization, its interaction with TATA-binding protein (TBP), and the expression of a set of cellular genes that are stimulated by IE2. On the contrary, WC5 targets the regulatory activity exerted by IE2 on different responsive viral promoters. Indeed, WC5 blocked the IE2-dependent negative regulation of the major immediate-early promoter by preventing IE2 binding to the crs element. Moreover, WC5 reduced the IE2-dependent transactivation of a series of indicator constructs driven by different portions of the early UL54 gene promoter, and it also inhibited the transactivation of the murine CMV early E1 promoter by the IE3 protein, the murine cytomegalovirus (MCMV) IE2 homolog. In conclusion, our results indicate that the overall anti-HCMV activity of WC5 depends on its ability to specifically interfere with the IE2-dependent regulation of viral promoters. Importantly, our results suggest that this mechanism is conserved in murine CMV, thus paving the way for further preclinical evaluation in an animal model.
INTRODUCTION
Human cytomegalovirus (HCMV) is one of the most common opportunistic viral pathogens in immunocompromised individuals and plays an important pathogenic role in chronic inflammatory diseases (1). Indeed, in transplant recipients, AIDS patients, and immunosuppressed individuals, HCMV represents an important cause of morbidity and mortality, as viral infection is associated with pneumonia, gastroenteritis, retinitis, and other life-threatening diseases (1). In addition, HCMV is the leading viral cause of congenital birth defects (1).
To date, to prevent and treat HCMV infections, there is no vaccine available, and only a limited number of drugs are licensed for treatment: ganciclovir (GCV), its oral prodrug valganciclovir, foscarnet, acyclovir, its prodrug valacyclovir, and cidofovir (2–4). The clinical utility of the current anti-HCMV drugs often has several drawbacks, such as an unfavorable safety profile characterized by severe acute and long-term toxicities and poor oral bioavailability that for some drugs requires intravenous administration (2). Moreover, no drugs have been approved for the treatment of congenital infection (5). The antiviral therapy is further complicated by the emergence of cross-resistant clinical strains, as the anti-HCMV compounds approved so far share a mechanism of action that targets the viral DNA polymerase (2). For all these reasons, there is still a strong need to develop new, safe, and effective antiviral compounds, possibly endowed with a new mechanism of action (6). In this regard, the identification of the viral factors that regulate the very early virus-host cell interactions as well as the functional characterization of the first viral proteins expressed in infected cells, such as the pivotal immediate-early 2 (IE2) protein, may now provide a rationale for the design of alternative antiviral strategies (7–9).
IE2 is an essential multifunctional protein that regulates crucial events in the HCMV replication cycle, such as viral early (E) gene activation, negative regulation of its own promoter, i.e., the major immediate-early promoter (MIEP), induction of host cell cycle progression, blocking of the cell cycle in S phase, and immunomodulation (10). IE2 is a 579-amino-acid (aa) protein able to dimerize, and its ability to self-interact has been related to its DNA-binding activity (10–12). The C-terminal region of IE2 (from aa 195 to 579) is required and sufficient for transcriptional activation, binding to DNA, dimerization, and protein-protein interactions (12).
A major role of IE2 is the activation of viral E gene expression, and this function is essential for the progression of the HCMV replication cycle, since it cannot be substituted by the activities of other viral or cellular proteins known so far (10). The DNA-binding activity of IE2 is required for maximal activation of the viral E promoters and thus for the subsequent steps in the HCMV lytic cycle (10, 13). Although cellular RNA polymerase II and TATA-binding protein (TBP) are recruited at viral E promoters in the absence of IE2, its presence at responsive promoters such as these is essential to achieve optimal E gene expression (13). Thus, it has been suggested that IE2 regulates E gene transcription both by direct binding to specific DNA sequences within responsive E gene promoters and by protein-protein interactions (10, 14).
When it accumulates in the infected cell, IE2 is also able to repress its own promoter, i.e., the MIEP, to avoid toxic effects related to its overexpression. This negative regulation occurs by direct binding of IE2 to a sequence called the cis repression signal (crs), which is located between the TATA box and the IE transcription start site (15–18). The crs regulatory element consists of a 14-bp A/T-rich region flanked on each side by two copies of a GC dinucleotide (19). The binding of IE2 to crs physically blocks the recruitment of RNA polymerase II and thus prevents the assembly of the preinitiation complex and subsequent IE gene transcription (20). Mutations in both crs and IE2 that impair the DNA-protein interaction have an abortive effect on virus replication (21). Furthermore, IE2 interacts with chromatin remodeling enzymes, such as histone deacetylases, to facilitate viral replication and regulate viral gene activation (22–25). In conclusion, IE2 is a master regulator of HCMV gene expression, and its activities are essential for the progression of the HCMV replication cycle (10, 26).
Moreover, there is compelling evidence that the IE2 protein plays a direct role in the pathogenesis of HCMV infection by inducing a radical change in host gene expression that leads to alterations in cell physiology and contributes to HCMV-induced cell cycle alterations, immunomodulation, and blocking of the inflammatory response (10, 27–29). For example, IE2 stimulates the expression of a set of genes involved in cell cycle progression, i.e., the E2F-responsive genes, thus providing HCMV with a mechanism to overcome cellular quiescence in infected cells. In fact, on one hand, IE2 promotes G0/G1 phase progression in infected quiescent cells, but on the other hand, in the same cells, it is also able to block the cell cycle progression by arresting them in the S phase, thus inhibiting cellular DNA synthesis in favor of viral DNA replication (10).
On the basis of this knowledge, it can be hypothesized that molecules able to decrease IE2 expression or inhibit the effects of its activity may be effective in blocking both HCMV replication and the virus-induced pathological phenomena at a very early stage of infection (7). In this scenario, inhibitors of IE2 might be particularly important for the treatment of patients who do not respond to the currently available inhibitors of viral DNA replication. To date, however, few molecules have been reported for their ability to inhibit IE2 expression and/or its functions (7, 8). In this regard, we have identified a 6-aminoquinolone (6-AQ) derivative termed WC5, endowed with a specific anti-HCMV activity (30, 31). WC5 and some analogues have been demonstrated to inhibit the IE2-induced transcriptional activation of two HCMV E gene promoters, i.e., UL112-113 and UL54, in a specific cell-based assay (31–33).
In the present study, we characterized further the mechanism of action of WC5 by focusing on its effects on IE2. All together, the results obtained suggest that the molecular mechanism whereby WC5 exerts anti-HCMV activity relies on its ability to specifically interfere with IE2-dependent regulation of viral responsive promoters. Other IE2 activities that we have examined were not affected by WC5. Interestingly, the inhibitory effect of WC5 was also observed in the context of the regulation of the murine CMV early E1 gene promoter by the immediate-early 3 protein (the murine cytomegalovirus [MCMV] homolog of HCMV IE2), thus providing a rationale for further development of WC5 in vivo.
MATERIALS AND METHODS
Compounds.
Compounds WC5 and WC5E were synthesized as previously described (30, 34) and solubilized in 100% dimethyl sulfoxide (DMSO). GCV (Cymevene; Roche) was obtained from a prescription pharmacy as a sodium salt.
Oligonucleotides.
All oligonucleotides used for PCR, mutagenesis, and sequencing, as well as the DNA probes for electrophoretic mobility shift assays (EMSAs), were obtained from Life Technologies.
Plasmids.
To create the pRSETB-IE2(290–579) plasmid, the sequence of IE2 coding aa 290 to 579 was amplified from the pSGIE86 plasmid (35) using primers IE2_290-579/FOR (5′-CAGTCAAGATCTTCGAGCCACCATGGGC-3′) and IE2_290-579/REV (5′-CACGTGGAATTCTTACTGAGACTTGTTCCTC-3′) and cloned into the BglII/EcoRI sites of the pRSETB plasmid (Life Technologies) downstream of the 6-histidine (6His) tag. To generate the pGEX-2T-IE2(290–579) plasmid, the IE2(290–579) fragment was subcloned from pRSETB-IE2(290–579) into the BamHI/EcoRI sites of the pGEX-2T plasmid (GE Healthcare Life Sciences) downstream of glutathione S-transferase (GST) coding sequence. To generate the pGEX-2T-IE55DN290 plasmid, the C-terminal portion of IE55 was amplified from the pSGIE55 plasmid (35) using primers IE2_290-579/FOR and IE2_290-579/REV and cloned into the BamHI/EcoRI sites of the pGEX-2T plasmid. To generate the pGEX-4T1-TBP plasmid, the TBP coding sequence was subcloned from the pCR-TBP2/3 plasmid (36), previously obtained by PCR amplification of the TBP sequence from the pKB104 plasmid (kindly provided by A. J. Berk, University of California, Los Angeles [UCLA], CA), into the BamHI/SalI sites of pGEX-4T1 (GE Healthcare Life Sciences) downstream of the GST coding sequence. To generate pD15-PA plasmid, the polymerase acidic (PA) coding sequence was amplified by PCR from the pcDNA-PA plasmid (from P. Digard, Roslin Institute, United Kingdom) using the PA-PD15/FOR and PA-PD15/REV primers (5′-TTTAAACTCGAGATGGAAGATTTTGTGCGAC-3′ and 5′-AAAAAAACGCGTCTAACTCAATGCATGTGT-3′, respectively) and cloned first into pCR2.1-TOPO vector (Life Technologies) and then the XhoI/MluI sites of pD15-UL44 vector (37). To create the pMIEP crs wild-type (wt) reporter plasmid, the HCMV IE promoter-enhancer sequence (positions −666 to +19 relative to the IE1/IE2 transcription initiation site [GenBank accession no. K03104.1) was amplified from the HCMV AD169 genome by PCR using the MIEP wild-type (wt) primer set (FOR, 5′-ACTGATGCTAGCGCATACGTTGTATCCATATC-3′, and REV, 5′-AGTGATAAGCTTGCGTCTCCAGGCGAT-3′; the underlined letters are the NheI and HindIII restriction sites used for cloning, respectively). The PCR fragments were cloned into the pGL3-basic vector (Promega) to obtain the pMIEP crs wt construct. To generate its derivative pMIEP crs mut (mutated), the crs element (from −14 to −1 with respect to the IE1/IE2 transcription initiation site) of pMIEP crs wt was changed, with the insertion of a unique restriction site (StuI, −13 to −7) with the QuikChange XL kit (Stratagene), using the crs mut oligonucleotide set (FOR, 5′-GTCTATATAAGCAGAGCTgaggcctgtccacGTCAGATCGCCTGGAGAC-3′, and REV, 5′-GTCTCCAGGCGATCTGACcatggacaggcctcTGCTCTGCTTATATAGAC-3′; the lowercase bold-type letters indicate mutated nucleotides), giving the pMIEP crs mut plasmid. Plasmids pSGIE86-P535A/Y537A and pSGIE86-H446A/H452A for eukaryotic expression of the corresponding mutant IE2 proteins were obtained by introducing desired point mutations in the pSGIE86 plasmid (35) with the QuikChange XL kit and the IE2 P535A/Y537A and IE2 H446A/H452A primer set, respectively (13). Plasmids pE(−207)Luc and pME(−207)Luc containing the firefly luciferase reporter gene under the control of wt and mutated cellular cyclin E promoters, respectively, were kindly provided by E. A. Thompson (Mayo Clinic Florida, FL) and were previously described (38). The pUL54-luciferase indicator plasmids pUL54-0.4, pUL54-0.3, pUL54-0.15, and pUL54-0.15 IR-1mut were generated as previously described (39). The pRL-SV40 vector expressing Renilla luciferase was from Promega. For the construction of the murine CMV (MCMV) pE1 reporter plasmid, pE1CAT (40) was digested on the BglII and HindIII restriction sites to obtain a 1,829-bp fragment that contains the MCMV E1 promoter responsive for IE3 transactivation activity (41). This fragment was cloned into the corresponding sites of the pGL3-basic vector (Promega) to obtain the pE1 construct. pIE3, which contains the coding sequence of MCMV IE3 gene, was described previously (40).
Recombinant protein expression and purification.
The recombinant 6His-IE2(290–579) protein (6His-IE2DN290) was purified from Escherichia coli BL21(DE3)pLysS strain harboring the pRSETB-IE2(290–579) plasmid, according to the protocol previously described for the 6His-PA protein (42). The recombinant proteins GST, GST-IE2D290, and GST-TBP were purified from E. coli BL21(DE3)pLysS harboring the pGEX-2T, pGEX-2T-IE2(290–579), and pGEX-4T1-TBP plasmids, respectively, according to the protocol previously described for GST-Ubc9 (43). The recombinant proteins GST-IE55DN290 and GST-PA were purified from E. coli BL21(DE3)pLysS harboring the pGEX-2T-IE55DN290 and pD15-PA plasmids, respectively, according to the protocol previously described for GST-UL44 (37).
GST pulldown assays.
For the GST pulldown assays, 0.03 nmol of either GST, GST-IE2DN290, GST-IE55DN290, GST-TBP, or GST-PA fusion protein was incubated for 2 h at 4°C with 0.6 nmol of 6His-IE2DN290 in a final volume of 0.1 ml in binding buffer (25 mM HEPES [pH 7.5], 12.5 mM MgCl2, 20% glycerol, 0.1% NP-40, 150 mM KCl, 0.15 mg/ml bovine serum albumin [BSA], and 1 mM dithiothreitol [DTT]) in the presence of either 0.2% DMSO, 50 μM WC5, or 50 μM WC5E. After incubation, the samples were loaded onto 0.1 ml of glutathione-Sepharose columns. The columns were then washed with 2 ml of NETN buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, and 0.5% NP-40), and fractions of 0.1 ml of the wash samples were collected (only samples derived from the last wash fraction were analyzed). The bound complexes were eluted with 0.3 ml of elution buffer (NETN plus 15 mM glutathione). The samples derived from the input, wash, and eluted fractions were then analyzed by Western blotting using a mouse monoclonal antibody (MAb) anti-6His tag (clone His-1, 1:1,000 dilution; Sigma). Immunocomplexes were detected with goat anti-mouse immunoglobulin Ab conjugated to horseradish peroxidase (HRP) (1:2,000 dilution; Santa Cruz Biotech) and visualized by enhanced chemiluminescence (LiteAblot Extend long-lasting chemiluminescent substrate kit; EuroClone).
Cells and virus.
Human embryonic kidney (HEK) 293 and U373-MG cells were grown in Dulbecco modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Life Technologies), 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate (P/S) (both from Life Technologies). NIH 3T3 cells were grown as monolayers in DMEM supplemented with 10% donor bovine serum (Life Technologies), 2 mM glutamine, and P/S. Low-passage human embryonic lung fibroblasts (HELFs) were grown in Eagle's minimal essential medium (MEM) (Life Technologies) supplemented with 10% FBS, 1 mM sodium pyruvate, 2 mM glutamine, and P/S. Quiescent HELF cells (arrested in G0/G1 phase) were obtained by culturing subconfluent monolayers for 1 week in medium containing 0.5% FBS (low-serum medium). Flow cytometry demonstrated that >90% of the cells were growth arrested. HCMV strain AD169 (VR-538) was purchased from the American Type Culture Collection (ATCC) (Manassas, VA).
Recombinant adenovirus vectors.
To create the pAC-CMV IE2 adenovirus shuttle vector, the pSGIE86 plasmid was digested with EcoRI and XbaI, and the cDNA of the HCMV IE2 protein was subsequently cloned into the corresponding sites of pAC-CMV (44). To generate a recombinant adenovirus expressing the HCMV IE2 protein (AdV-IE2), the pAC-CMV IE2 and pJM17 vectors (Microbix Biosystems) were cotransfected into subconfluent HEK 293 cells using the CaPO4-DNA coprecipitation method. Recombinant AdV-IE2 was then identified by immunoblotting. After several rounds of plaque purification, recombinant AdV-IE2 was amplified on HEK 293 cells. A recombinant adenovirus expressing the E. coli β-galactosidase gene (AdV-LacZ) was used as a control (45).
Immunoblotting.
Whole-cell protein extracts of AdV-IE2-infected cells were prepared as previously described (46), separated by 10% SDS-PAGE, and then analyzed by Western blotting with a mouse anti-HCMV IE antigen MAb (clone E13; Argene Biosoft) and a mouse anti-actin MAb (1:2,000 dilution; Chemicon International) used as a control for protein loading. Immunocomplexes were detected with a sheep anti-mouse immunoglobulin Ab conjugated to HRP (1:2,000 dilution; Amersham).
Cell transfections and adenoviral transductions.
For the transfection experiments with U373-MG cells, the cells were seeded in 24-well plates and the next day were transiently cotransfected using calcium phosphate (CellPhect transfection kit; GE Healthcare) with 0.05 μg of pMIEP crs wt or mut plasmid and, where indicated, 1 μg of wt or mutated pSGIE86 plasmid in a 1:5 ratio, as well as 0.05 μg of pRL-SV40 plasmid as a control for transfection efficiency. The total DNA amount was equalized with pSG5 empty vector (Promega). After incubation for 4 h at 37°C, the transfection mixtures were removed, and medium containing either 50 μM WC5 or WC5E, or DMSO (0.2%) as a control, was added to the cells. At 48 h posttransfection, the cells were harvested, and both the firefly luciferase and Renilla luciferase activities were measured by a luciferase assay system and a Renilla luciferase assay system (Promega), according to the manufacturer's instructions. For the transfection/transduction experiments with HELF cells, they were grown on 24-well plates and cotransfected using SuperFect reagent (Qiagen) with 0.9 μg of each pUL54-derived plasmid and with 0.1 μg of pRL-SV40 plasmid as an internal control. At 24 h posttransfection, the cells were infected with either AdV-IE2 or AdV-LacZ at a multiplicity of infection (MOI) of 10 for 2 h at 37°C and then treated with 50 μM WC5, 50 μM WC5E, or 0.2% DMSO as a control. At 48 h postransduction, the cells were harvested, and the firefly luciferase and Renilla luciferase activities were measured. For all the experiments, the values were normalized by dividing the values obtained for firefly luciferase by the values obtained for Renilla luciferase and expressed as relative luciferase units (RLU).
Real-time RT-PCR.
Quiescent HELFs were infected with HCMV (MOI, 0.5 PFU/cell) or with AdV-IE2 or AdV-LacZ (MOI, 10 PFU/cell), and, where indicated, they were treated 2 h postinfection (p.i.) with 50 μM WC5 or WC5E. At 48 h p.i., the total cellular RNA was extracted with the NucleoSpin RNA kit (Macherey-Nagel). One microgram of RNA was then retrotranscribed using the RevertAid H Minus FirstStrand cDNA synthesis kit (Fermentas) in a final volume of 20 μl. Two microliters of cDNAs (or water as a control) was amplified in duplicate by quantitative PCR (qPCR) using the Brilliant SYBR green QPCR master mix (Stratagene) in a final volume of 25 μl. The sequences of the oligonucleotides used for assessing mRNA levels were as follows: TS (FOR, 5′-GCAAAGAGTGATTGACACCATCAA-3′, and REV, 5′-CAGAGGAAGATCTCTTGGATTCCAA-3′), CDK2 (FOR, 5′-GCTAGCAGACTTTGGACTAGCCAG-3′, and REV, 5′-AGCTCGGTACCACAGGGTCA-3′), RR1 (FOR, 5′-GGAGGAATTGGTGTTGCTGT-3′, and REV, 5′-GCTGCTCTTCCTTTCCTGTG), and β-actin (FOR, 5′-CAAAAGCCTTCATACATCTC-3′, and REV, 5′-TCATGTTTGAGACCTTCAA-3′) as an internal control gene for the normalization data. Following an initial denaturing step at 95°C for 2 min to activate 0.75 units of Platinum Taq DNA polymerase (Invitrogen), the cDNAs were amplified for 30 cycles of 95°C for 1 min, 58°C for 1 min, and 72°C for 1 min. For quantitative analysis, the log change in fluorescence was plotted against the cycle number, and a threshold was set for the changes in fluorescence at a point in the linear PCR amplification phase (threshold cycle [CT]). The CT values for each gene were normalized to the CT values for β-actin using the ΔCT equation. The level of target RNA, normalized to the endogenous β-actin reference and relative to the 12-h infected cells, was calculated by the comparative CT method and the 2−ΔΔCT equation.
Electrophoretic mobility shift assays.
To generate the crs-derived probes, single-stranded forward and reverse oligonucleotides containing wt crs sequences (FOR, 5′-GCTGAGCTCGTTTAGTGAACCGTCA-3′, and REV, 5′-GATCTGACGGTTCACTAAACGAGCT-3′; the italic uppercase characters indicate the wt crs sequence) or mutated (mut) crs sequences (FOR, 5′-GCTGAGCTgagggcctgtccaCGTCA-3′, and REV, 5′-GATCTGACGtggacaggcctcAGCT-3′; the mutated nucleotides in mut crs are lowercase bold-type letters, and the italic uppercase characters indicate the wt crs sequence) were annealed by incubating 500 pmol of each complementary strand in 10 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, and 50 mM NaCl for 5 min at 95°C, followed by gradual cooling at room temperature (RT). For the EMSAs, 2.5 μg of purified 6His-IE2DN290 was incubated at 20°C for 20 min in a final volume of 20 μl in Tris-EDTA (TE) buffer (pH 8.0) containing 10% glycerol, 3 μg/μl BSA, 1 mM MgCl2, 12.5 ng/μl poly(dA)-oligo(dT), 5 ng/μl sonicated and denatured salmon sperm DNA (Sigma), 0.1 mM DTT, 100 mM KCl, and 25 pmol of the double-stranded crs-derived probes. Test 6-AQs (0.5 mM) were added to the reaction products where indicated, and control reactions with 0.2% DMSO were included. After incubation, the reaction mixtures were separated on a native 5% polyacrylamide gel in 0.5× TBE buffer (90 mM Tris-borate, 2 mM EDTA [pH 8.0]) at 4°C, followed by staining with GelRed (Biotium, Inc.), according to the manufacturer's protocol. For EMSAs using the sequence derived from the cellular cyclin E promoter, the same protocol was used but with the following annealed oligonucleotide sequences for cellular cyclin E (CcnE): FOR, 5′-AGCCGGCGCGGCCGCCAGCGCGGTGT-3′, and REV, 5′-ACACCGCGCTGGCGGCCGCGCCGGCT-3′.
Statistical analysis.
All statistical tests were performed using the GraphPad Prism software. The data are presented as the means ± standard deviations (SD). The data were analyzed for significance using a paired t test and were considered statistically significant at a P value of ≤0.05.
RESULTS
WC5 does not affect IE2 dimerization and its interaction with TBP.
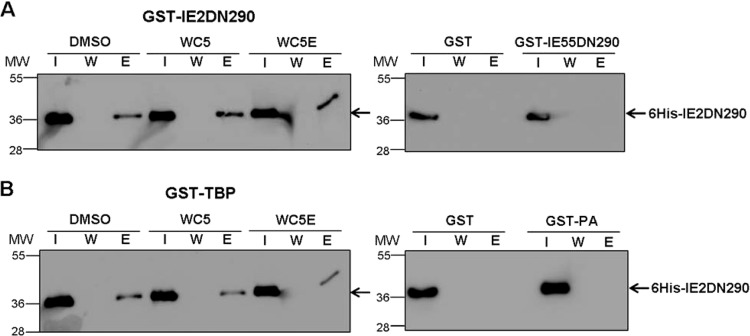
We previously observed that WC5 interferes with the transactivating activity of IE2 on two HCMV E gene promoters (31). Since IE2 regulates gene expression by both protein-protein interactions and direct binding to specific DNA sequences within E-responsive promoters (10), we first wished to determine whether the inhibitory effect of WC5 might be due to interference with two protein-protein interactions that are required for the transactivating activity of IE2, such as the interaction with itself and with cellular TBP. To this end, the GST pulldown assays were set up with a truncated form of IE2, IE2DN290, which has been shown to be sufficient for both dimerization and interaction with TBP (12, 47) and which was fused to either to GST or a 6His tag. The purified recombinant GST-IE2DN290 and 6His-IE2DN290 proteins were mixed, incubated in the absence or presence of WC5 or its inactive derivative WC5E as a control (30), and then tested for dimerization by the GST pulldown assays. As shown in Fig. 1A (left), the dimerization between the two differently tagged versions of the C-terminal segment of IE2 occurred independently of the presence of WC5 or WC5E, since bands corresponding to 6His-IE2DN290 were detected in the eluted fraction of each reaction mixture. A control reaction in which 6His-IE2DN290 was incubated with either GST alone or GST fused to a truncated form of IE2 that is dimerization defective, i.e., GST-IE55DN290 (11), ruled out the possibility that the presence of 6His-IE2DN290 in the eluted samples was due to nonspecific interactions (Fig. 1A, right).
FIG 1.
WC5 does not affect IE2 protein-protein interactions. (A) GST pulldown assays performed with 6His-IE2DN290 and GST-IE2DN290 recombinant proteins in the absence or presence of WC5 or WC5E (left). As a control of specificity, reactions with either GST alone or GST-IE55DN290 were also included (right). (B) GST pulldown assays performed with 6His-IE2DN290 and GST-TBP in the absence or presence of either WC5 or WC5E (left). As a control of specificity, reactions with either GST alone or GST-PA were also included (right). The input (I), the last wash fraction (W), and eluted (E) proteins of each reaction mixture were visualized by Western blotting with an anti-6His antibody. The positions of the molecular mass markers (in kilodaltons) are indicated on the left side of each panel.
Since the interaction of IE2 with TBP has been reported to be critical for its transactivating activity (48), we next investigated whether WC5 and WC5E may affect the interaction between 6His-IE2DN290 and a GST-TBP fusion protein. As shown in Fig. 1B (left), both compounds did not exert any effect on the interaction between 6His-IE2DN290 and GST-TBP. Furthermore, to confirm that the presence of 6His-IE2DN290 in the eluted samples was due to specific interaction with TBP, we included control reactions with either GST alone or an unrelated GST fusion protein, such as GST-PA, which contains the PA protein of influenza virus. As shown in Fig. 1B (right), no interaction was detected under these conditions. Taken together, these results indicate that WC5 does not affect the ability of IE2 to interact with itself or with TBP, thus excluding the possibility that its inhibitory activity is due to interference with these protein-protein interactions.
WC5 does not affect the IE2-dependent stimulation of some cellular responsive genes.
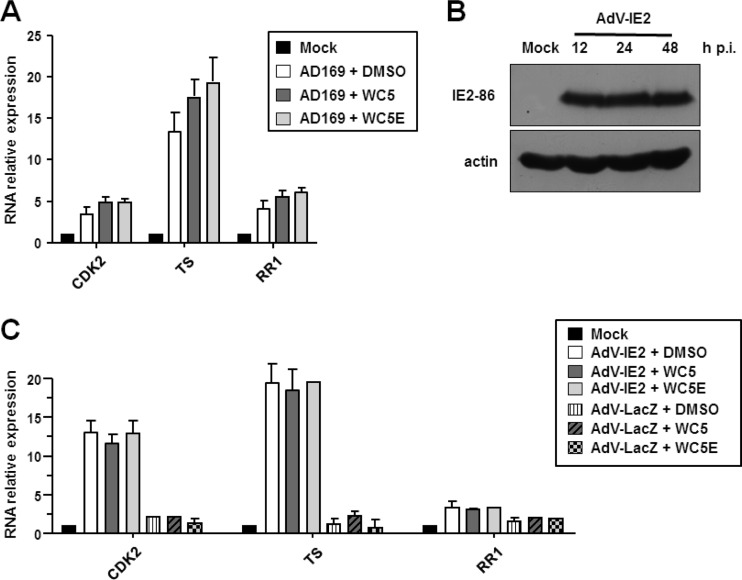
The observation that WC5 does not inhibit IE2 dimerization and its interaction with TBP prompted us to investigate whether WC5 might interfere with the protein-protein interactions that are related to IE2-dependent regulation of cellular gene expression. Thus, we tested the effect of WC5 on the expression of a set of selected E2F-dependent cellular genes whose transcription has been reported to be stimulated either by HCMV infection or upon expression of IE2 alone and that are thought to be regulated by IE2 via protein-protein interactions rather than by the direct binding of IE2 to their promoters (49, 50). To this end, the mRNA levels of the genes encoding cellular thymidylate synthetase (TS), cyclin-dependent kinase 2 (CDK2), and ribonucleotide reductase 1 (RR1) were measured by real-time reverse transcription-PCR (RT-PCR) in quiescent HELFs that had been infected with HCMV for 48 h in the absence or presence of WC5 or its inactive derivative WC5E as a control. As shown in Fig. 2A, HCMV infection, as is known, increased the mRNA levels of TS, CDK2, and RR1. However, WC5 did not significantly affect the accumulation of these HCMV-stimulated cellular mRNAs.
FIG 2.
WC5 does not affect the HCMV- or IE2-dependent stimulation of cellular E2F-responsive genes. (A) Growth-arrested HELFs were infected with HCMV at an MOI of 0.5 PFU/cell or were mock infected (mock) and, where indicated, treated with WC5 or WC5E. Total cellular RNA was isolated at 48 h p.i. and reverse transcribed. qPCR was then carried out with the appropriate primers for cyclin-dependent kinase 2 (CDK2), thymidylate synthetase (TS), ribonucleotide reductase 1 (RR1), and β-actin (as a control). Next, the RNA levels were normalized according to the expression of the β-actin gene. (B) Expression of the IE2 protein in HELFs transduced with a recombinant AdV-IE2. The HELFs were infected with AdV-IE2 (MOI, 10 PFU/cell) or mock infected (mock). At the indicated times p.i., the total cell extracts were analyzed by immunoblotting with an anti-IE2 antibody. β-Actin immunodetected with an MAb served as an internal control. (C) Effects of WC5 or WC5E on cellular E2F-responsive genes in IE2-expressing quiescent HELFs. The total RNA was isolated at 48 h p.i. from quiescent HELFs that had not been infected (mock) or had been infected with AdV-IE2 or AdV-LacZ at an MOI of 10 PFU/cell and treated as described above. The RNA levels were then analyzed by qPCR and normalized to β-actin as an endogenous control. (A and C) Reported RNA relative expression levels are normalized to the values of the mock-infected cells, which were set at a value of 1. The data shown are the means ± SD from three independent experiments.
Next, we evaluated the effect of WC5 on the expression of the abovementioned HCMV-responsive genes when stimulated by IE2 alone. Quiescent HELFs were infected with either an IE2-expressing adenoviral vector (AdV-IE2) or a β-galactosidase-expressing adenovirus (AdV-LacZ) as a control. The successful expression of transduced exogenous IE2 protein was assessed by immunoblotting analysis (Fig. 2B). IE2- or LacZ-expressing HELFs were then treated with WC5 or WC5E, and at 48 h after drug exposure, the mRNA levels of the target genes were measured by real-time RT-PCR. As expected, in the AdV-IE2-transduced cells, a significant increase in TS, CDK2, and RR1 mRNA levels was measured, which was not observed in the cells transduced with the control AdV-LacZ (Fig. 2C). In keeping with the results obtained in the HCMV-infected HELFs, the increase in mRNA levels stimulated by IE2 overexpression was not affected by WC5 or WC5E (Fig. 2C).
Taken together, these results indicate that WC5 does not affect the IE2-mediated expression of representative IE2-responsive cellular genes.
WC5 abrogates the IE2-dependent repression of HCMV MIEP.
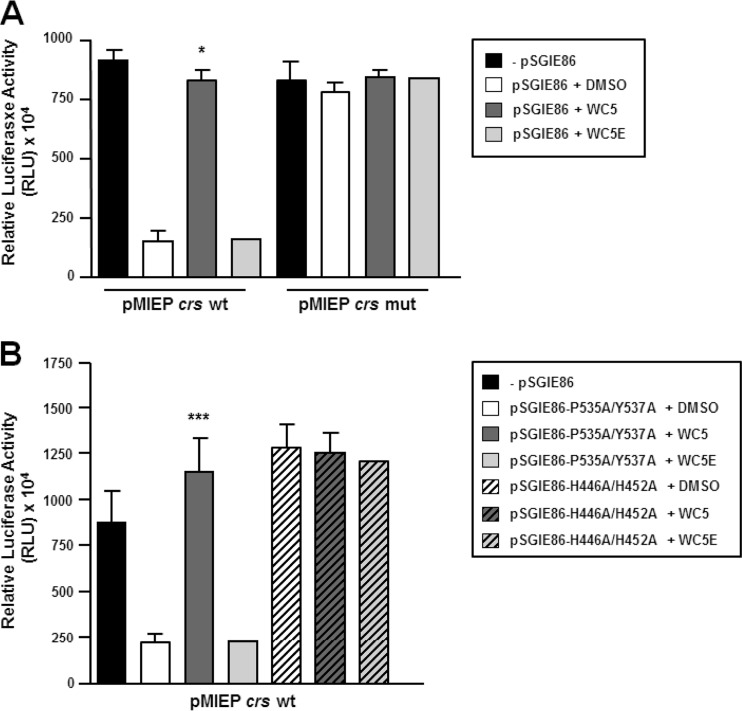
The observation that WC5 does not tamper with the IE2-dependent gene regulation via protein-protein interactions led us to examine whether the DNA binding of IE2 to a responsive promoter might represent a target of WC5. To this end, we took into account the IE2-dependent repression of its own promoter, i.e., the MIEP, which depends on the direct interaction between IE2 and the crs sequence (10). To investigate the effect of WC5 on the IE2-mediated negative regulation of MIEP, permissive U373-MG cells were cotransfected with a plasmid containing the luciferase reporter gene under the control of MIEPs bearing either the wt or a mutated (mut) crs sequence (designated pMIEP crs wt and pMIEP crs mut, respectively) and with a plasmid expressing wt IE2 (pSGIE86). As expected (13), IE2 expression caused a ∼5-fold reduction in MIEP transcriptional activity in cells cotransfected with pMIEP crs wt (Fig. 3A). The lack of a significant reduction in luciferase expression in the extracts prepared from cells that had been cotransfected with the pMIEP crs mut plasmid confirmed the specificity of the observed IE2-dependent repression (Fig. 3A). However, when the transfected cells were treated with WC5, the luciferase expression was restored to levels comparable to those observed in the absence of IE2 expression only in cells transfected with wt crs-containing plasmid but not in cells transfected with pMIEP crs mut (Fig. 3A). As expected, the inactive WC5E did not exert any effect on IE2-mediated repression of MIEP (Fig. 3A). Moreover, the effect of WC5 on MIEP repression by IE2 was dose dependent (see Fig. S1 in the supplemental material available at http://www.medicinamolecolare.unipd.it/sites/dipartimenti.it/files/Supplemental%20Material.pdf), thus further indicating a specific effect for IE2-crs interaction. To confirm these results, we tested two mutated versions of the IE2 protein for their ability to repress the MIEP in the presence of WC5. The first mutant protein, bearing the P535A/Y537A substitutions, is competent for MIEP repression but is no longer able to transactivate viral E gene promoters (13). The second mutant, with H446A/H452A substitutions, is not able to repress the MIEP or transactivate viral E promoters (13). As shown in Fig. 3B, the repressive activity of IE2 P535A/Y537A on MIEP was inhibited by WC5 but not by the inactive analog WC5E. In contrast, no effect of WC5 or WC5E was observed in the cells cotransfected with MIEP crs wt and with a plasmid expressing the IE2 H446A/H452A protein (Fig. 3B). Thus, these results further indicate that WC5 specifically interferes with the IE2-dependent repression of MIEP.
FIG 3.
WC5 abrogates the IE2-mediated repression of MIEP. (A) U373-MG cells were transfected with a luciferase reporter plasmid containing HCMV MIEP (pMIEP-crs) with either wild-type (wt) or mutated (mut) crs or cotransfected with an IE2-expressing plasmid (pSGIE86) and then treated with 0.2% DMSO or 50 μM WC5 or WC5E. (B) U373-MG cells were cotransfected with the MIEP-crs wt plasmid and plasmids expressing mutant P535A/Y537A and H446A/H452A IE2 proteins and then treated with 0.2% DMSO or 50 μM WC5 or WC5E. (A and B) Transfection mixtures also contained a plasmid constitutively expressing Renilla luciferase to normalize variations in transfection efficiency. The reported values represent the means ± SD from three independent experiments in duplicate and are expressed as relative luciferase units (RLU) (i.e., light units of firefly luciferase per 104 Renilla luciferase light units as determined at 48 h posttransfection); *, P < 0.05, and ***, P < 0.001, versus calibrator sample (pMIEP crs wt or mut plus pSGIE86 wt or mut plus DMSO).
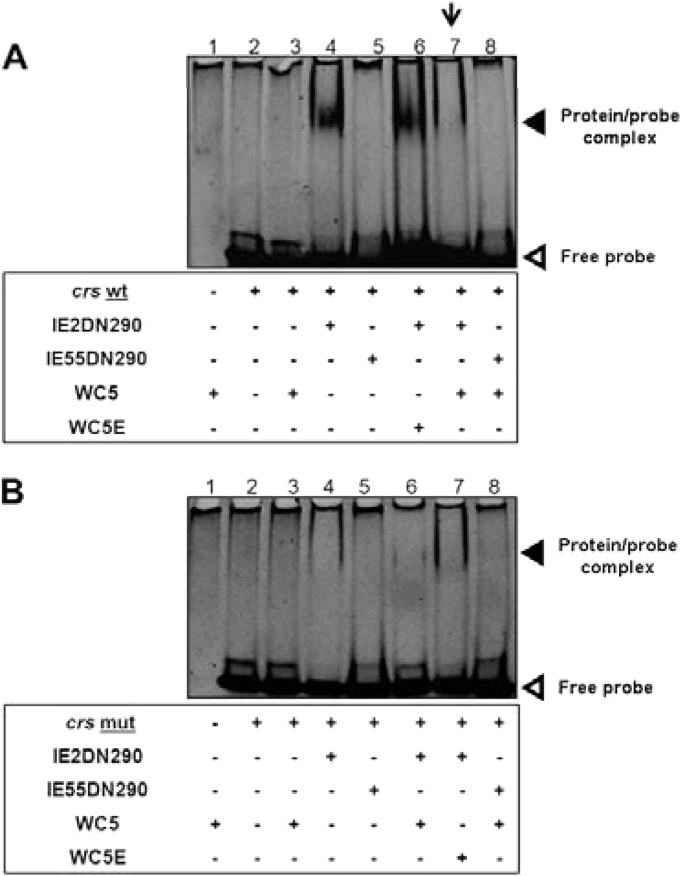
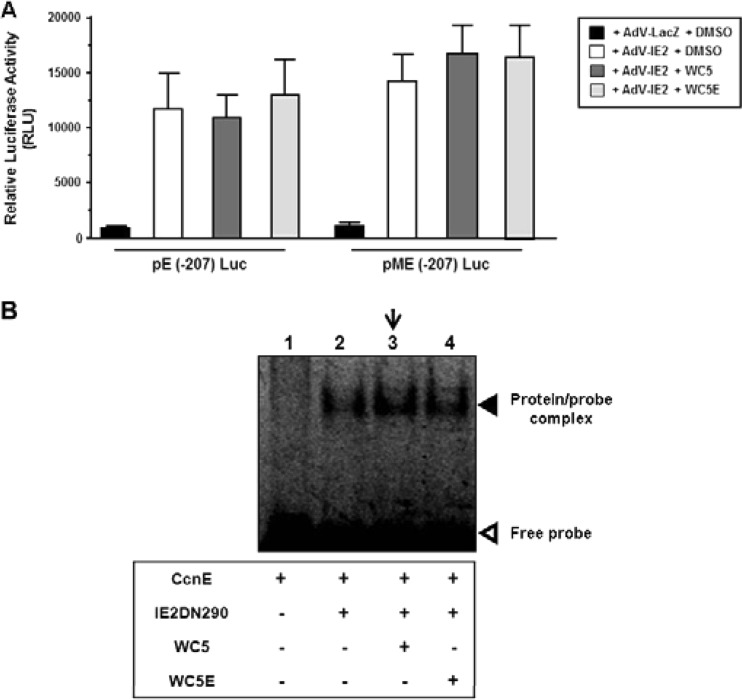
To support further this observation, we then analyzed the effect of WC5 and of WC5E as a control on the ability of the C-terminal fragment of IE2 (aa 290 to 579), which has been shown to be sufficient for DNA binding (10, 12, 47, 51), to interact in vitro with a wt crs-derived probe. The IE2 C-terminal segment was expressed as the recombinant 6His-IE2DN290 protein. As shown in Fig. 4A, in the EMSAs, a shifted protein-probe complex was observed when the 6His-IE2DN290 protein was incubated with wt crs (lane 4). This complex was not detected when a mutated crs was used as a probe, thus demonstrating the specificity of IE2 binding (Fig. 4B, lane 4). The addition of WC5 to either wt or mut crs alone did not produce any effect on the mobility of the probes (compare lane 2 to lane 3 in both Fig. 4A and B). In contrast, the presence of WC5 in the reaction mixture containing 6His-IE2DN290 and wt crs prevented the formation of the protein-probe shifted complex (Fig. 4A, lane 7), thus indicating that WC5 is able to interfere with the binding of IE2 to wt crs. The inability of WC5E to interfere with the formation of the IE2DN290-crs complex confirmed that the inhibition exerted by WC5 was specific (Fig. 4A, lane 6). As further specificity controls, EMSAs with the DNA-binding defective GST-IE55DN290 protein (11) and either wt or mut crs probes were performed. In these samples, the protein-probe complex was not detected independently of the presence of WC5 (lane 5 in both Fig. 4A and B).
FIG 4.
WC5 inhibits the binding of IE2 to the MIEP crs wt element. Electrophoretic mobility shift assays (EMSAs) were performed to evaluate the effects of WC5 and WC5E on the IE2DN290-crs interaction by incubating the 6His-IE2DN290 protein with MIEP crs-containing probes either wt (A) or mutated (mut) (B) in the absence or presence of WC5 and WC5E. The binding reaction products were then resolved on 5% native polyacrylamide gels and stained with GelRed. The down-facing arrow indicates the absence of the protein-probe complex in the presence of WC5.
Taken together, the results of this section indicated that WC5 abrogates the IE2-mediated repression of MIEP and prevents the binding of IE2 to the crs element. Thus, in the context of the HCMV replicative cycle, WC5, besides inhibiting the IE2-dependent transactivation of two E promoters, i.e., the UL54 and UL112-113 promoters (31), also interferes with another essential function of IE2, such as the negative regulation of MIEP.
A proximal 150-bp segment is sufficient to mediate the WC5-dependent inhibition of HCMV UL54 gene promoter transactivation.
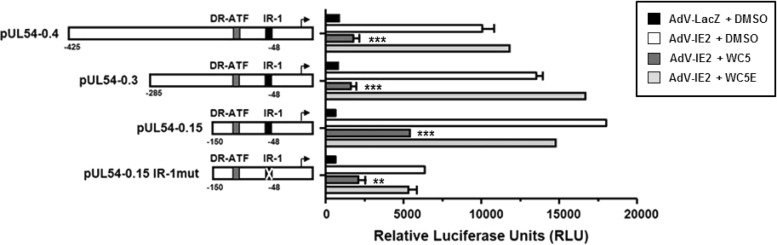
We previously observed that WC5 is able to inhibit the IE2-dependent transactivation of two E genes of HCMV, i.e., UL112-113 and UL54, both in transfected cells and in the context of viral infection (31). Thus, to gain further insights into the mechanism of this inhibitory activity, we wished to identify the minimal portion of an E gene promoter activated by IE2 still sensitive to WC5 inhibition. To this end, the promoter of the UL54 gene, an essential HCMV gene that encodes the DNA polymerase catalytic subunit, was chosen as a prototypic E gene promoter. Luciferase reporter plasmids containing UL54 gene promoter segments (pUL54) with progressive 5′ deletions (thus leaving approximately 0.4, 0.3, and 0.15 kbp upstream from the transcription start site; Fig. 5) were therefore generated (39). The HELF cells were transfected with the different pUL54 constructs and then infected with either AdV-IE2 or AdV-LacZ as a control. After transduction with recombinant AdVs, the cells were finally incubated in the absence or presence of WC5 or WC5E. As shown in Fig. 5, IE2 expression transactivated all three pUL54-derived constructs, although at different levels: from about 12-fold for pUL54-0.4 up to >30-fold for pUL54-0.15. Treatment with WC5 but not WC5E reduced the transcriptional activities of all the tested reporter constructs, including the shortest pUL54-0.15 construct, >3-fold.
FIG 5.
A 150-bp segment of the UL54 gene promoter is sufficient to mediate the inhibitory effect of WC5 on IE2-dependent transactivation. HELFs were transfected with luciferase reporter plasmids containing segments of HCMV UL54 promoter progressively deleted from the 5′ end and/or mutated in the IR-1 element and subsequently infected with AdV-IE2 or AdV-LacZ at an MOI of 10 PFU/cell. Where indicated, the cells were treated with 0.2% DMSO, WC5, or WC5E. At 48 h p.i., luciferase expression was determined. The transcriptional activity of each indicator construct is expressed in relative luciferase units (RLU). The data shown are the means ± standard error of the mean from three independent experiments; **, P < 0.01, and ***, P < 0.001, versus the calibrator sample (AdV-IE2 plus DMSO).
The UL54 gene promoter contains an 8-bp inverted repeat element 1 (IR-1) located between −54 and −43 nucleotides (nt) relative to the transcription start site, which has been shown to be required for both HCMV- and IE2-mediated transactivation (52, 53). IE2 and the cellular transcription factor Sp1 have been found in an IR-1 DNA-protein complex detected in IE2-overexpressing cells (54). In addition, it was demonstrated that IR-1 contains an unconventional binding site for the cellular transcription factor Sp1, which seems to play a role in UL54 gene promoter basal activity in both IE2-transfected cells and HCMV-infected cells (54, 55). To investigate the involvement of the IR-1 element in WC5-mediated inhibition of IE2 transactivation, a luciferase reporter construct driven by the minimal UL54 promoter mutated in the IR-1 element (pUL54-0.15 IR-1mut) was generated and examined in the transfection/transduction experiments. As shown in Fig. 5, and in accordance with previous observations (54, 55), inactivation of the IR-1 element caused about a 3-fold decrease in the IE2-mediated UL54 promoter transactivation compared to that of pUL54-0.15, which confirmed the prominent role of IR-1 in the regulation of the overall UL54 gene promoter response to IE2 (52). Despite the lack of a functional IR-1 element, IE2 was still able to transactivate the pUL54-0.15 IR-1mut construct about 9-fold, and this IE2-dependent transactivation was reduced by WC5 treatment by >60% (Fig. 5). Similar results were obtained in another experimental setting, i.e., U373-MG cells that had been transfected with the pUL54 indicator constructs together with an IE2-expressing plasmid (pSGIE86) and then treated with WC5 and WC5E (data not shown).
All together, these results indicate that a segment of 150 bp upstream from the UL54 transcription initiation site is sufficient to mediate the inhibitory activity of WC5 on the IE2-dependent transactivation of the UL54 promoter, and that WC5 retains its ability to downregulate IE2-mediated transactivation in the absence of a functional IR-1 element.
WC5 does not affect the IE2-dependent activation of cellular cyclin E promoter.
The ability of WC5 to interfere with the binding of IE2 to viral promoters led us to investigate whether the transactivation of a cellular gene regulated by IE2 via direct binding to its promoter, such as cyclin E, might be a target of WC5. Cellular cyclin E (CcnE) is upregulated upon HCMV infection in quiescent cells to promote G0/G1 transition (56), and its expression can be directly stimulated by IE2 (38). To assess the effect of WC5 on the IE2-mediated transactivation of the CcnE promoter, we transfected HELFs with reporter constructs containing either a portion of the wt CcnE promoter, as in pE(−207)Luc (which includes nucleotides −207 to +77 of the human CcnE promoter), or a mutated version wherein the binding sites for the cellular transcription factor E2F at −16 and +7 have been mutated, as in pME(−207)Luc (38). The transfected cells were then infected with AdV-IE2 or AdV-LacZ as a control and treated with WC5 or WC5E. As shown in Fig. 6A, the expression of the IE2 protein transactivated the wt CcnE promoter, and the disruption of E2F binding sites in pME(−207)Luc did not affect the IE2-dependent transactivation of the cyclin E promoter, as previously reported (38). The addition of WC5 did not produce any effect on the IE2-dependent transactivation of the CcnE promoter independently of the presence of mutations in the E2F binding sites (Fig. 6A). Similar results were obtained with U373-MG cells that had been transfected with the CcnE promoter-derived reporter plasmids together with an IE2-expressing plasmid and then treated with WC5 or WC5E (data not shown). In addition, we investigated the effect of WC5 on the binding of the 6His-IE2DN290 protein to the CcnE promoter-derived probe corresponding to +35 to +60 nucleotides that were previously reported to interact with IE2 (38). As expected, a probe-protein complex was observed when 6His-IE2DN290 was incubated with a CcnE probe (Fig. 6B, lane 2). However, according to the results obtained from the transfection experiments, WC5 and WC5E did not affect the formation of the shifted protein-probe complex (Fig. 6B, lanes 3 and 4). Together, these data indicate that WC5 does not interfere with the binding of IE2 to the CcnE promoter.
FIG 6.
WC5 does not affect the binding of IE2 to the cellular cyclin E promoter. (A) HELF cells were transfected with a luciferase reporter plasmid containing either the wt [pE(−207)Luc] or mutated [pME(−207)Luc] cellular cyclin E promoter and subsequently infected with AdV-IE2 or AdV-LacZ at an MOI of 10 PFU/cell. The cells were treated with 0.2% DMSO or 50 μM WC5 or WC5E. At 48 h p.i., luciferase expression was determined and expressed as relative luciferase units (RLU). The reported values represent the means ± SD from three independent experiments in duplicate. (B) EMSA performed to evaluate the effects of WC5 and WC5E on the interaction of IE2DN290 with a cyclin E promoter-derived probe (CcnE). The down-facing arrow indicates the absence of any effect of WC5 on the mobility of the protein-probe complex compared to Fig. 4A, lane 7.
WC5 inhibits the IE3-dependent transactivation of an E gene of murine CMV.
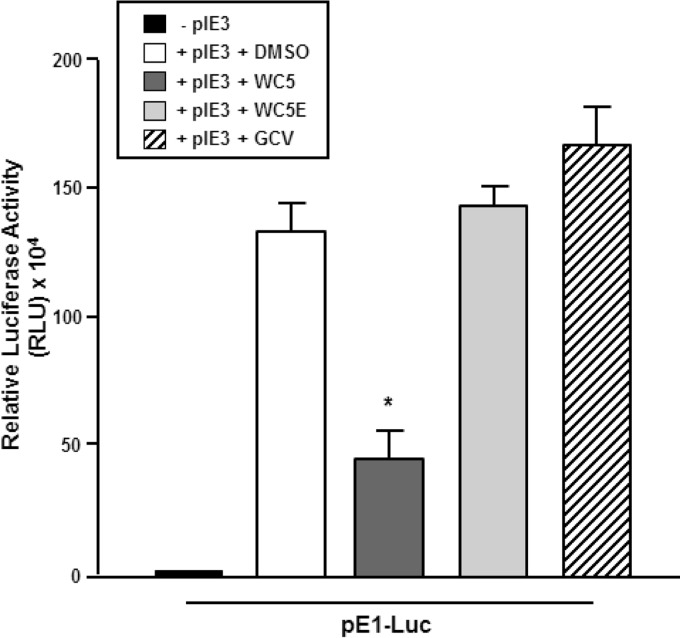
We previously reported that WC5 was active against murine cytomegalovirus (MCMV) replication in vitro (30). Since MCMV represents a suitable model for in vivo studies for validating the potential use in the prevention and/or control of HCMV infection of in vitro-selected anti-HCMV agents (57), we next investigated whether WC5 may also affect the transcriptional activation of an E gene of MCMV. The MCMV genome encodes an HCMV IE2 homolog, the IE3 protein, which has been shown to be required for the transactivation of viral E and L genes and thus for the progression of the MCMV replicative cycle (40, 58–60). To analyze the effects of WC5 on IE3-mediated E gene transactivation, an indicator construct driven by the promoter of the prototype early E1 gene of MCMV (pE1-Luc) was generated and transfected into NIH 3T3 cells, together with an IE3-expressing plasmid in the absence or presence of WC5 or GCV. As shown in Fig. 7, the expression of the IE3 protein strongly transactivated the E1 promoter. WC5 treatment inhibited the reporter gene expression about 3-fold compared to that of the untreated samples. The absence of an inhibitory effect in the transfected cells treated with either the inactive analog WC5E or GCV indicated the specificity of the WC5 effect on the IE3-mediated transactivation of pE1.
FIG 7.
WC5 inhibits the IE3-dependent transactivation of the early E1 gene promoter of MCMV. NIH 3T3 cells were transfected with a plasmid containing luciferase reporter gene under the control of the E1 promoter (pE1-Luc) or cotransfected with an IE3-expressing plasmid (pIE3) and then treated with 0.2% DMSO, 50 μM WC5, WC5E, or GCV. The reported values represent the means ± SD from three independent experiments in duplicate and are expressed as RLU. *, P < 0.05 versus calibrator sample (pIE3 plus DMSO).
These results suggest that the mechanism of action of WC5 against both HCMV and MCMV might be conserved and, importantly, demonstrates that MCMV infection is a valid preclinical model for the study of the antiviral activity of WC5 in vivo.
DISCUSSION
The currently available anti-HCMV drugs have several drawbacks; in addition, they cannot prevent the reactivation of latent viral infection and are not approved for the treatment of congenital infections. The identification of novel anti-HCMV agents that can block IE gene expression and/or IE functions at very early stages without causing major adverse effects may provide an alternative strategy for inhibiting HCMV reactivation, replication, and immunopathogenesis (7, 8). The feasibility of this approach has been validated by fomivirsen, a 21-base phosphorothioate oligodeoxynucleotide complementary to IE2 mRNA and thus able to block IE2 protein expression, which has been approved for intraocular application in patients with HCMV retinitis (2).
To contribute to the characterization and development of new IE2-targeting molecules, this study was undertaken to investigate further the mechanism of action of the 6-aminoquinolone WC5 against HCMV IE2. We previously demonstrated that WC5 interferes with the IE2-mediated transactivation of two HCMV E gene promoters, i.e., the UL112-113 and UL54 promoters, in a specific cell-based assay (31). Here, we have examined whether other functions of IE2 might be affected by WC5 by focusing on some known interactions of IE2 with DNA and proteins that contribute to viral replication. We demonstrated that WC5 interferes with some functions of IE2 that are essential for productive HCMV replication, i.e., the autoregulation of MIEP and the transactivation of viral E genes that are required for the progression of the HCMV replicative cycle (10, 21), while it has no effect on other IE2 activities (e.g., the upregulation of some cellular genes).
First, we analyzed the effects of WC5 on those IE2 protein-protein interactions that have been shown to be relevant for its autoregulatory activity and are involved in the IE2-mediated transactivation of viral E promoters, such as dimerization or interaction with TBP. IE2 interacts with itself (10, 12) and with the C-terminal basic repeat domain of TBP (10, 14) through domains that are overlapping and located within the C-terminal segment of the protein. The same domains are required also for transactivation and DNA binding (10, 61). The results of the GST pulldown assays demonstrated that WC5 does not affect the interaction of IE2 with itself or with TBP in vitro (Fig. 1), suggesting that its antiviral activity most likely does not stem from an interference with these IE2 protein-protein interactions. This conclusion is further sustained by the lack of any significant inhibitory activity of WC5 on the expression of a set of E2F-responsive cellular genes which can be stimulated by either HCMV infection or IE2 expression (49) and which are thought to be indirectly regulated via protein-protein interactions rather than direct binding of IE2 to their promoters. In fact, the treatment with WC5 of quiescent cells either infected with HCMV or transduced with a recombinant IE2-expressing AdV did not affect the stimulation of cellular CDK2, RR1, and TS mRNAs (Fig. 2).
DNA-protein interactions are a traditional target of quinolone antimicrobial agents (62); therefore, we next examined the effect of WC5 on the most well-characterized interaction of IE2 with a DNA element, namely, its binding to the crs of HCMV MIEP that leads to the repression of IE gene expression (10). WC5 showed the ability to abrogate the repression of MIEP exerted by the wt IE2 protein, as well as by an IE2 mutant still able to repress the MIEP but lacking transactivating activity (13). The results of the EMSAs indicated that WC5 prevents the formation of an IE2-crs complex (Fig. 4). Thus, it can be concluded that the abrogation of the IE2-dependent repression of MIEP in transfected cells treated with WC5 is due to interference with the binding of the protein to the crs element. The 3-carboxylic acid moiety in the quinolone scaffold of WC5 has been implicated in quinolone/nucleic acid binding; thus, it might be involved in the interaction with the crs, since the analog WC5E, which lacks this group due to esterification, does not inhibit either HCMV replication (30) or the binding of IE2 to a crs-derived probe (Fig. 4).
Another HCMV promoter regulated by IE2 is that of the essential gene UL54. We previously reported that WC5 interferes in transfected cells with the IE2-dependent transactivation of a reporter construct bearing the entire promoter of UL54 (31). Here, we have identified a minimal portion of the UL54 gene promoter sufficient to mediate the sensitivity to WC5 in a segment of 150 bp upstream from the transcription start site. In fact, according to previous studies, this portion of the UL54 5′-flanking region mediates the IE2-dependent transactivation of the whole gene promoter at a level comparable to that of the entire promoter (52). Although the regulation of the UL54 promoter by IE2 has been the subject of previous studies (52–54), the only cis-acting element in the promoter with an established role in IE2-dependent transactivation that has been characterized so far is the IR-1 sequence (54, 55). However, a direct binding of IE2 to the UL54 promoter has not been detected, although IE2 and the cellular transcription factor Sp1 have been found as components of an IR-1 DNA-protein complex that had been detected in extracts prepared from IE2-overexpressing U373-MG cells (55). It was observed that the binding of Sp1 to the IR-1 element was strongly increased in the presence of IE2, thus suggesting that IE2 may induce a functional modulation of Sp1 that enables it to bind IR-1, and that the association of IE2 with the IR-1 response element might occur through this cellular factor (55). The inhibitory activity of WC5 on the transcriptional activity of pUL54-0.15 might thus stem from interference with the formation of a transcriptional competent IR-1 DNA-protein complex. In a previous study, we observed that WC5 did not exert any effect on two different viral promoters that require Sp1 for their basal transcriptional activity, thus suggesting that a direct interference of WC5 with the binding of Sp1 to DNA is unlikely (30). Sp1 is also involved in the activation of the intercellular adhesion molecule-1 (ICAM-1) promoter by IE2, and IE2 augments the transcriptional activity of the ICAM-1 promoter in concert with pp71 (63). However, when the Sp1 site within the ICAM-1 promoter was mutated, a residual (but lower) promoter activity was still observed, thus suggesting that functional Sp1 binding sites are not strictly essential for the IE2-mediated transactivation of the ICAM-1 promoter. Here, a similar residual transactivation potential was observed with the pUL54-0.15 IR-1mut construct, wherein the IR-1 element was inactivated (Fig. 5). The inhibitory effect of WC5 on the minimal pUL54 construct bearing a mutated IR-1 that was measured as well (Fig. 5) might thus suggest that outside the IR-1 sequence, an additional IE2-responsive DNA region(s) might be present. Further studies will be required to identify possible additional IE2-responsive elements, if there are any.
Among those IE2 functions that require an interaction with DNA regulatory elements, the inhibitory effect of WC5 seems to be confined specifically to the regulation of viral elements, such as MIEP and E gene promoters. In fact, we have observed that in cells transfected with indicator constructs containing the promoter of cellular cyclin E, WC5 does not significantly affect gene reporter activity (Fig. 6A). The expression of the cyclin E gene is greatly stimulated upon HCMV infection or IE2 expression (38, 49). Moreover, it was observed that IE2 directly interacts with the cyclin E promoter, thus promoting its transactivation (38). In our EMSAs, WC5 did not interfere with the binding of IE2 to a probe derived from the cyclin E promoter (Fig. 6B), thus indicating that not all IE2 functions that implicate direct or indirect binding to DNA are targeted by WC5. Differences in the sequences of the IE2-binding sites, their localization within the promoters, the possible DNA secondary structures that might originate after IE2 binding, and IE2 interactions with specific cellular chromatin remodeling enzymes might account for the differential effects of WC5 on IE2-responsive promoters. Once the crystallographic structure of the IE2 bound to DNA is available, this and other questions can be addressed.
The preference of WC5 for CMV promoters that emerged from this study is in agreement with the specific antiviral activity demonstrated by this compound against HCMV over other herpesviruses, such as herpes simplex virus 1 (HSV-1), and other viruses, such as HIV-1, and also with the absence of any effect on the transactivation of promoters of two constitutive cellular genes and of HIV-1 long terminal repeat (LTR) (30, 34). Furthermore, the inhibitory activity of WC5 is specific for IE2 over IE1, the other major protein product of the MIE gene locus that shares the first 85 amino acids with IE2. In fact, we observed that in transient assays, WC5 did not interfere with the transactivation of the thymidylate synthetase gene promoter, a specific IE1-responsive promoter (50) (see Fig. S2 in the supplemental material available at http://www.medicinamolecolare.unipd.it/sites/dipartimenti.it/files/Supplemental%20Material.pdf).
In conclusion, we have identified a first mechanism of action of WC5 against HCMV in its ability to interfere with the interaction of IE2 to crs within the MIEP, thus blocking its negative regulation. The negative autoregulation of the MIEP by IE2 is essential for efficient HCMV replication, since recombinant viruses that fail to repress the MIEP have not been isolated (10, 21). A possible indirect effect of WC5 on the IE2-dependent epigenetic mechanisms that contribute to MIEP regulation cannot be excluded. In fact, IE2-mediated negative regulation of the MIEP also depends on its ability to recruit repressive chromatin remodeling enzymes (23); thus, the interference with IE2 binding to crs exerted by WC5 might have an additional detrimental effect on the correct silencing of the MIEP and the progression of the HCMV cycle. A second mechanism of action of WC5 against HCMV entails the inhibition of IE2-dependent transactivation of viral E gene promoters that are required for viral DNA replication and the progression of the HCMV replication cycle, such as the UL54 and UL112-113 promoters. Therefore, the inhibitory effect of WC5 on these two different IE2-dependent activities contributes to the overall anti-HCMV activity.
WC5 may thus be an attractive candidate for a new class of anti-HCMV drugs that exert their effects via novel pathways that target IE functions. Our in vitro studies warrant further investigations to evaluate whether WC5 treatment may result in antiviral activity in animal models of acute infection, as well as in reactivation from latency. Indeed, our experiments demonstrated that WC5 is active against MCMV replication (30) and prevents the transactivation of a prototypic MCMV E gene mediated by the IE3 protein, the murine homolog of HCMV IE2. In fact, MCMV IE3 shares a striking homology with IE2 not only at the amino acid sequence level (the C-terminal 200 amino acids are identical, and an additional 18 are conserved) but also in their regulatory functions; indeed, IE3 is essential for virus growth, it exerts repressive activity on its own ie1/ie3 promoter, and it is sufficient to activate MCMV E gene expression (40, 60, 64). Given this high similarity between HCMV IE2 and MCMV IE3, it is reasonable that the inhibitory activity of WC5 is conserved, thus making possible the investigation of the therapeutic potential of WC5 in the murine model of cytomegalovirus infection.
ACKNOWLEDGMENTS
We thank Adriana Mattiolo, Federica Tolio, and Matteo Bronzini for experimental help and technical assistance. We also thank E. A. Thompson for providing the cyclin E promoter-containing plasmids, A. J. Berk for the pKB104 plasmid, P. Digard for the pcDNA-PA plasmid, L. F. Johnson for the phTS −243/+30 construct, and M. Messerle for the IE3- and pE1-containing plasmids.
This work was supported by the Ministero dell'Istruzione, Università e Ricerca (MURST EX60%, Progetto di Ricerca di Ateneo 2007 CPDA074945, and PRIN 20085FF4J4 [to A.L.] and MURST EX60% and PRIN 2010-11 2010PHT9NF [to G.G.]), by the University of Padova (Progetto Strategico 2008 [to G.P.]), and by ESCMID (ESCMID grant 2013 [to B.M.]).
Footnotes
Published ahead of print 25 August 2014
REFERENCES
- 1.Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417–470. 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber A, Harter G, Schubert A, Bunjes D, Mertens T, Michel D. 2009. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 10:191–209. 10.1517/14656560802678138. [DOI] [PubMed] [Google Scholar]
- 3.Prichard MN, Kern ER. 2011. The search for new therapies for human cytomegalovirus infections. Virus Res. 157:212–221. 10.1016/j.virusres.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emery VC. 2012. Cytomegalovirus: recent progress in understanding pathogenesis and control. QJM 105:401–405. 10.1093/qjmed/hcr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. 2013. The “silent” global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 26:86–102. 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lischka P, Zimmermann H. 2008. Antiviral strategies to combat cytomegalovirus infections in transplant recipients. Curr. Opin. Pharmacol. 8:541–548. 10.1016/j.coph.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Scholz M, Doerr HW, Cinatl J. 2001. Inhibition of cytomegalovirus immediate early gene expression: a therapeutic option? Antiviral Res. 49:129–145. 10.1016/S0166-3542(01)00126-7. [DOI] [PubMed] [Google Scholar]
- 8.Mercorelli B, Lembo D, Palù G, Loregian A. 2011. Early inhibitors of human cytomegalovirus: state-of-art and therapeutic perspectives. Pharmacol. Ther. 131:309–329. 10.1016/j.pharmthera.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercorelli B, Gribaudo G, Palù G, Loregian A. 2014. Approaches for the generation of new anti-cytomegalovirus agents: identification of protein-protein interaction inhibitors and compounds against the HCMV IE2 protein. Methods Mol. Biol. 1119:349–363. 10.1007/978-1-62703-788-4_18. [DOI] [PubMed] [Google Scholar]
- 10.Stinski MF, Petrik DT. 2008. Functional roles of the human cytomegalovirus essential IE86 protein. Curr. Top. Microbiol. Immunol. 325:133–152. 10.1007/978-3-540-77349-8_8. [DOI] [PubMed] [Google Scholar]
- 11.Furnari BA, Poma E, Kowalik TF, Huong SM, Huang ES. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou CJ, Zong J, Waheed I, Hayward GS. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrik DT, Schmitt KP, Stinski MF. 2007. The autoregulatory and transactivating functions of the human cytomegalovirus IE86 protein use independent mechanisms for promoter binding. J. Virol. 81:5807–5818. 10.1128/JVI.02437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukac DM, Harel NY, Tanese N, Alwine JC. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pizzorno MC, O'Hare P, Sha L, LaFemina RL, Hayward GS. 1988. Trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermiston TW, Malone CL, Stinski MF. 1990. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J. Virol. 64:3532–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pizzorno MC, Hayward GS. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Hermiston TW, Stinski MF. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waheed AA, Ablan SD, Sowder RC, Roser JD, Schaffner CP, Chertova E, Freed EO. 2010. Effect of mutations in the human immunodeficiency virus type 1 protease on cleavage of the gp41 cytoplasmic tail. J. Virol. 84:3121–3126. 10.1128/JVI.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Jupp R, Stenberg RM, Nelson JA, Ghazal P. 1993. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J. Virol. 67:7547–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isomura H, Stinski MF, Kudoh A, Nakayama S, Murata T, Sato Y, Iwahori S, Tsurumi T. 2008. A cis element between the TATA box and the transcription start site of the major immediate-early promoter of human cytomegalovirus determines efficiency of viral replication. J. Virol. 82:849–858. 10.1128/JVI.01593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevels M, Paulus C, Shenk T. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. U. S. A. 101:17234–17239. 10.1073/pnas.0407933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeves M, Murphy J, Greaves R, Fairley J, Brehm A, Sinclair J. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998–10009. 10.1128/JVI.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JJ, Kim YE, Pham HT, Kim ET, Chung YH, Ahn JH. 2007. Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J. Gen. Virol. 88:3214–3223. 10.1099/vir.0.83171-0. [DOI] [PubMed] [Google Scholar]
- 25.Cuevas-Bennett C, Shenk T. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525–9536. 10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870–1878. 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor RT, Bresnahan WA. 2005. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J. Virol. 79:3873–3877. 10.1128/JVI.79.6.3873-3877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor RT, Bresnahan WA. 2006. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression. J. Virol. 80:10763–10771. 10.1128/JVI.01195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor RT, Bresnahan WA. 2006. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 80:920–928. 10.1128/JVI.80.2.920-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercorelli B, Muratore G, Sinigalia E, Tabarrini O, Biasolo MA, Cecchetti V, Palù G, Loregian A. 2009. A 6-aminoquinolone compound, WC5, with potent and selective anti-human cytomegalovirus activity. Antimicrob. Agents Chemother. 53:312–315. 10.1128/AAC.00988-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loregian A, Mercorelli B, Muratore G, Sinigalia E, Pagni S, Massari S, Gribaudo G, Gatto B, Palumbo M, Tabarrini O, Cecchetti V, Palù G. 2010. The 6-aminoquinolone WC5 inhibits human cytomegalovirus replication at an early stage by interfering with the transactivating activity of viral immediate-early 2 protein. Antimicrob. Agents Chemother. 54:1930–1940. 10.1128/AAC.01730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massari S, Mercorelli B, Sancineto L, Sabatini S, Cecchetti V, Gribaudo G, Palù G, Pannecouque C, Loregian A, Tabarrini O. 2013. Design, synthesis and evaluation of WC5 analogues as inhibitors of human cytomegalovirus immediate-early 2 protein, a promising target for anti-HCMV treatment. ChemMedChem 8:1403–1414. 10.1002/cmdc.201300106. [DOI] [PubMed] [Google Scholar]
- 33.Luganini A, Caposio P, Mondini M, Landolfo S, Gribaudo G. 2008. New cell-based indicator assays for the detection of human cytomegalovirus infection and screening of inhibitors of viral immediate-early 2 protein activity. J. Appl. Microbiol. 105:1791–1801. 10.1111/j.1365-2672.2008.03927.x. [DOI] [PubMed] [Google Scholar]
- 34.Cecchetti V, Parolin C, Moro S, Pecere T, Filipponi E, Calistri A, Tabarrini O, Gatto B, Palumbo M, Fravolini A, Palù G. 2000. 6-Aminoquinolones as new potential anti-HIV agents. J. Med. Chem. 43:3799–3802. 10.1021/jm9903390. [DOI] [PubMed] [Google Scholar]
- 35.Klucher KM, Sommer M, Kadonaga JT, Spector DH. 1993. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol. Cell. Biol. 13:1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loregian A, Bortolozzo K, Boso S, Sapino B, Betti M, Biasolo MA, Caputo A, Palù G. 2003. The Sp1 transcription factor does not directly interact with the HIV-1 Tat protein. J. Cell. Physiol. 196:251–257. 10.1002/jcp.10271. [DOI] [PubMed] [Google Scholar]
- 37.Loregian A, Appleton BA, Hogle JM, Coen DM. 2004. Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J. Virol. 78:158–167. 10.1128/JVI.78.1.158-167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bresnahan WA, Albrecht T, Thompson EA. 1998. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J. Biol. Chem. 273:22075–22082. 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 39.Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 8:e1002498. 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messerle M, Buhler B, Keil GM, Koszinowski UH. 1992. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J. Virol. 66:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arai Y, Ishiwata M, Baba S, Kawasaki H, Kosugi I, Li RY, Tsuchida T, Miura K, Tsutsui Y. 2003. Neuron-specific activation of murine cytomegalovirus early gene e1 promoter in transgenic mice. Am. J. Pathol. 163:643–652. 10.1016/S0002-9440(10)63691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muratore G, Goracci L, Mercorelli B, Foeglein Á, Digard P, Cruciani G, Palù G, Loregian A. 2012. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc. Natl. Acad. Sci. U. S. A. 109:6247–6252. 10.1073/pnas.1119817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong L, Kamitani T, Fujise K, Caskey LS, Yeh ET. 1997. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272:28198–28201. 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 44.Becker TC, Noel RJ, Coats WS, Gómez-Foix AM, Alam T, Gerard RD, Newgard CB. 1994. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 43:161–189. 10.1016/S0091-679X(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 45.Caposio P, Gugliesi F, Zannetti C, Sponza S, Mondini M, Medico E, Hiscott J, Young HA, Gribaudo G, Gariglio M, Landolfo S. 2007. A novel role of the interferon-inducible protein IFI16 as inducer of proinflammatory molecules in endothelial cells. J. Biol. Chem. 282:33515–33529. 10.1074/jbc.M701846200. [DOI] [PubMed] [Google Scholar]
- 46.Luganini A, Caposio P, Landolfo S, Gribaudo G. 2008. Phosphorothioate-modified oligodeoxynucleotides inhibit human cytomegalovirus replication by blocking virus entry. Antimicrob. Agents Chemother. 52:1111–1120. 10.1128/AAC.00987-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macias MP, Stinski MF. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc. Natl. Acad. Sci. U. S. A. 90:707–711. 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris SM, Bullock B, Westgard E, Zhu H, Stenberg RM, Kerry JA. 2010. Functional properties of the human cytomegalovirus IE86 protein required for transcriptional regulation and virus replication. J. Virol. 84:8839–8848. 10.1128/JVI.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song YJ, Stinski MF. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99:2836–2841. 10.1073/pnas.052010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gribaudo G, Riera L, Rudge TL, Caposio P, Johnson LF, Landolfo S. 2002. Human cytomegalovirus infection induces cellular thymidylate synthase gene expression in quiescent fibroblasts. J. Gen. Virol. 83(Pt 12):2983–2993. [DOI] [PubMed] [Google Scholar]
- 51.Stinski MF, Isomura H. 2008. Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency. Med. Microbiol. Immunol. 197:223–231. 10.1007/s00430-007-0069-7. [DOI] [PubMed] [Google Scholar]
- 52.Kerry JA, Priddy MA, Stenberg RM. 1994. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J. Virol. 68:4167–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerry JA, Priddy MA, Jervey TY, Kohler CP, Staley TL, Vanson CD, Jones TR, Iskenderian AC, Anders DG, Stenberg RM. 1996. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J. Virol. 70:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, O'Neill J, Barbosa MS. 1998. Transcription factor Sp1 mediates cell-specific trans-activation of the human cytomegalovirus DNA polymerase gene promoter by immediate-early protein IE86 in glioblastoma U373MG cells. J. Virol. 72:236–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luu P, Flores O. 1997. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J. Virol. 71:6683–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalejta RF, Shenk T. 2002. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 7:d295–d306. [DOI] [PubMed] [Google Scholar]
- 57.Kern ER. 2006. Pivotal role of animal models in the development of new therapies for cytomegalovirus infections. Antiviral Res. 71:164–171. 10.1016/j.antiviral.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 58.Keil GM, Ebeling-Keil A, Koszinowski UH. 1987. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J. Virol. 61:526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angulo A, Ghazal P, Messerle M. 2000. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129–11136. 10.1128/JVI.74.23.11129-11136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martínez FP, Cosme RSC, Tang Q. 2010. Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation. J. Gen. Virol. 91:2664–2676. 10.1099/vir.0.022301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lang D, Stamminger T. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richter S, Parolin C, Palumbo M, Palù G. 2004. Antiviral properties of quinolone-based drugs. Curr. Drug Targets Infect. Disord. 4:111–116. 10.2174/1568005043340920. [DOI] [PubMed] [Google Scholar]
- 63.Kronschnabl M, Stamminger T. 2003. Synergistic induction of intercellular adhesion molecule-1 by the human cytomegalovirus transactivators IE2p86 and pp71 is mediated via an Sp1-binding site. J. Gen. Virol. 84:61–73. 10.1099/vir.0.18703-0. [DOI] [PubMed] [Google Scholar]
- 64.Bühler B, Keil GM, Weiland F, Koszinowski UH. 1990. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J. Virol. 64:1907–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]