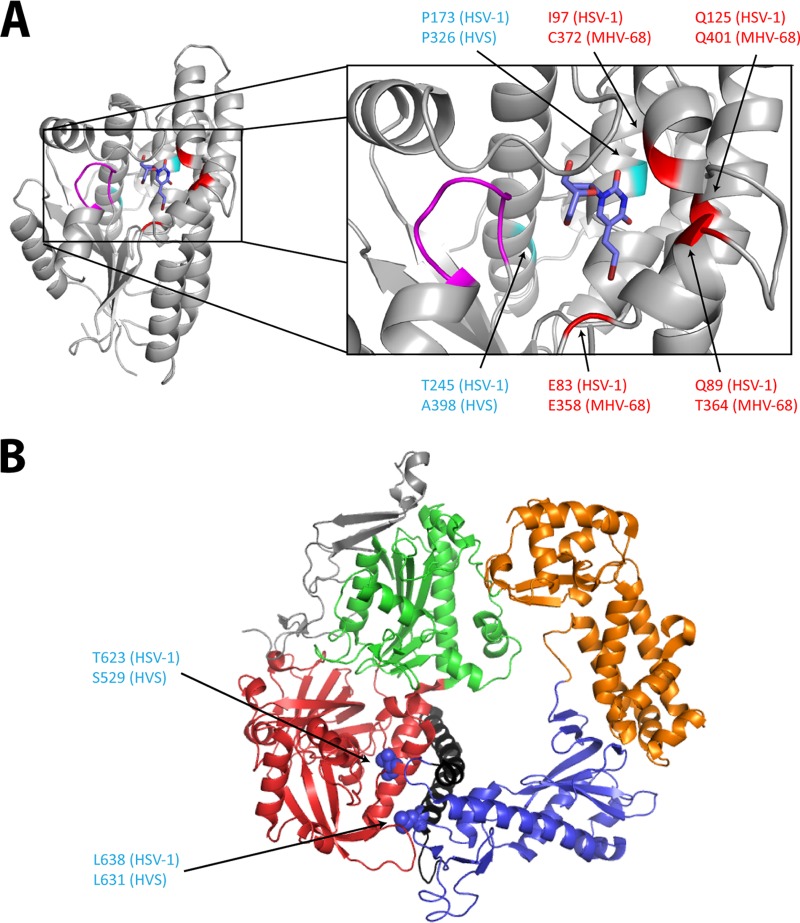
FIG 6.
Visualization of the mutations identified in HVS and MHV-68, at the homologous positions of HSV-1 TK and DNA polymerase three-dimensional (3D) structures. (A) BVDU (blue) is bound to the phosphate acceptor site of the HSV-1 TK (PDB code 1KI8). The p-loop (magenta) motif present in all the ATP-binding proteins is important for the interactions with the β and γ phosphates of the phosphate donor. The positions of the mutations identified in HVS are shown in cyan, and those identified in MHV-68 are in red. Amino acid changes at positions C372 (MHV-68) and A398 (HVS) described in this study were found under selection with BVDU. The other amino acid changes (shown in red at position P326 in HVS and positions Q401, E358, and T364 in MHV-68) were identified after selection with a novel pyrimidine nucleoside derivative (i.e., HDVD) possessing high levels of selectivity and potency against gammaherpesviruses. All the identified mutations are present in the first shell of residues surrounding the substrate (here BVDU) in the active site. (B) 3D structure of the HSV-1 DNA polymerase, showing the different domains: pre-NH2 domain (gray), NH2-terminal domain (red), 3′-5′-exonuclease domain (green), polymerase palm subdomain (blue), finger subdomain (black), and thumb subdomain (orange). Amino acid changes at the homologous position (indicated in blue) are present in the NH2-terminal domain and the finger subdomain. All the pictures were generated using PyMol graphic system (open source; v0.99rc9).