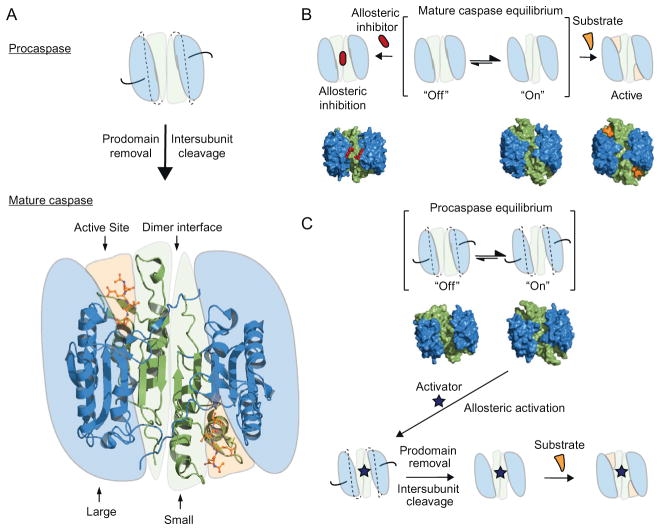
Figure 8.5.
Mechanism of caspase regulation. (A) Executioner procaspase maturation requires proteolysis of the N-terminal prodomain (black lines) and cleavage of the inter-subunit linker (dashed lines) between the large and small subunits. The crystal structure of mature caspase-7 is shown (PDB ID 1F1J) highlighting the dimer interface, the large and small subunits and the Ac-DEVD-CHO peptide occupying the active site (shown in sticks). (B) Mechanism of allosteric inhibition of caspases, showing the hotspot for allosteric binding located at the dimer interface. The surface representation of experimentally determined X-ray structures of caspase-7 is shown below the cartoons. From left to right, the allosteric-site ligand-bound structure of caspase-7 complexed with DICA, the ligand-free apo caspase-7 structure in an “on” conformation, and the active-site ligand-bound structure of caspase-7 in complex with the Ac-DEVD-CHO peptide (PDB ID 1SHJ, 1K86, 1F1J, respectively). (C) Mechanism of allosteric activation of procaspases. The brackets denote the open and closed active-site equilibrium, with the surface of the X-ray structures of procaspase-3 in the “on” and “off” states (PDB ID 4JR0 and 4JQY, respectively). Upon binding of a hypothetical allosteric activator, the proenzyme is locked into an “on” conformation, allowing possible proteolytic cleavage of the prodomain and intersubunit linkers.