Figure 3. The LAVA element and evidence for LAVA-mediated early transcription termination.
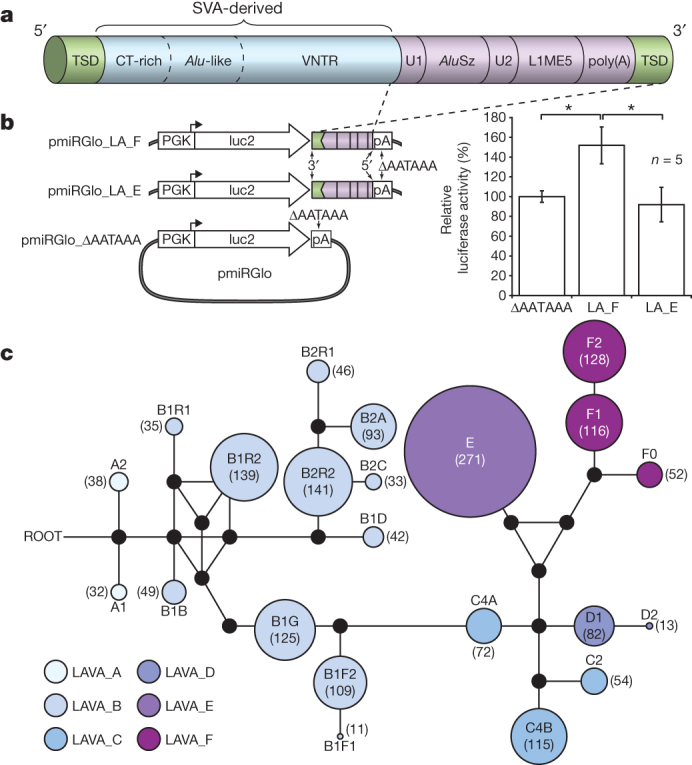
a, Schematic view of the LAVA element highlights the main components that originated from common repeats (L1, Alu, VNTR and Alu-like). Target-site duplications (TSDs) and the poly(A) tail are also indicated. b, Luciferase reporter constructs used to assay for LAVA-mediated early transcriptional termination (left panel) and results of the luciferase reporter assay (right panel) showing increased luciferase activity by ∼50% relative to the background for pmiRGlo_LA_F (*P = 0.0013) (see Supplementary Information section S7.8) n = 5, five biological replicates, from five independent transfections done for each experimental condition tested. The experiment shown was replicated twice in the laboratory. Statistics were carried out using a Student’s t-test (two sided), P values for all pairwise comparisons LA_F vs. LA_E, ΔPA vs. LA_F, and ΔPA vs. LA_E respectively (with 95% CI) were adjusted for multiple comparisons according to the Bonferroni method. Centre values show the average, error bars indicate standard deviation. c, A median-joining network showing the relationships among the 22 LAVA subfamilies generated by comparing the 3′ intact LAVA elements. Coloured circles represent subfamilies and their size is proportional to the number of elements in the subfamily (numbers inside each circle). Black dots represent hypothetical sequences connecting adjacent subfamilies. All possible relationships are shown. Branch lengths are not drawn to scale.
