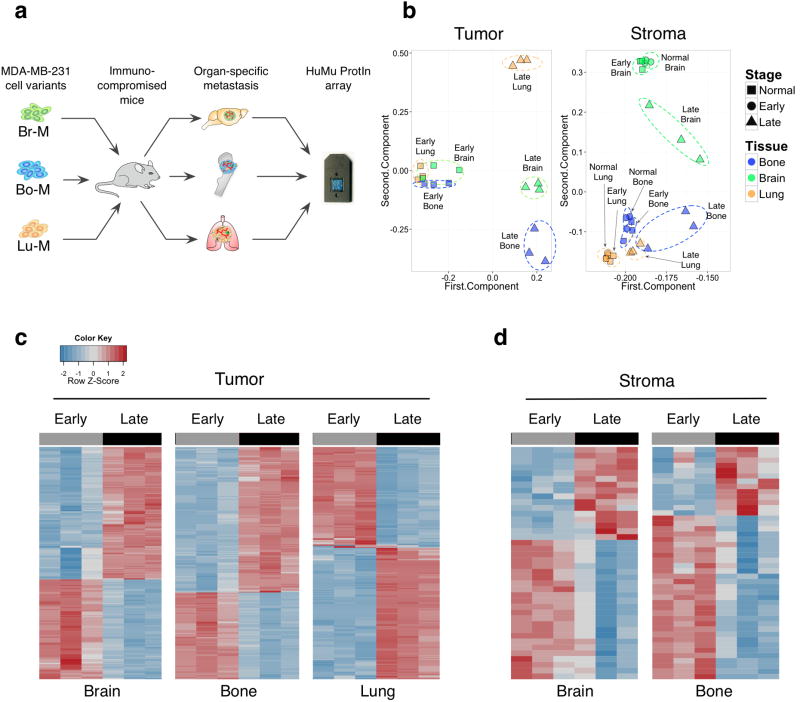
Figure 1. HuMu ProtIn array enables simultaneous acquisition of gene expression changes in tumor and stromal cells.
(a) Schematic of the experimental design employed to analyze tumor stroma interactions in different metastatic microenvironments using the dual species-specific HuMu ProtIn (Human/ Murine Proteases and Inhibitors) array in xenografted animals. Br-M = brain metastatic, Bo-M = bone metastatic, and Lu-M = lung metastatic variants of the MDA-MB-231 cell line. (b) Principal component analysis of the HuMu array data: the 1st and 2nd component are plotted on the x and y axes respectively. These two components together represent the largest sources of variation in the dataset. The first and second components represent 89.98% and 8.44% respectively of the variance in the tumor gene space, and 90.83% and 4.06% in the stromal gene space. This analysis revealed variation in tumor gene expression that was predominantly associated with differences between early- and late-stage metastasis. Meanwhile, variation in the stroma was associated with both stage and tissue. Dotted ellipses were drawn manually to indicate related data points within stage or organ. (c,d) Heatmaps of (c) tumor- and (d) stroma-derived genes that were differentially expressed between early and late metastases across different organ sites. The lung stroma did not show extensive differences between early and late stages (Supplementary Table 1g).