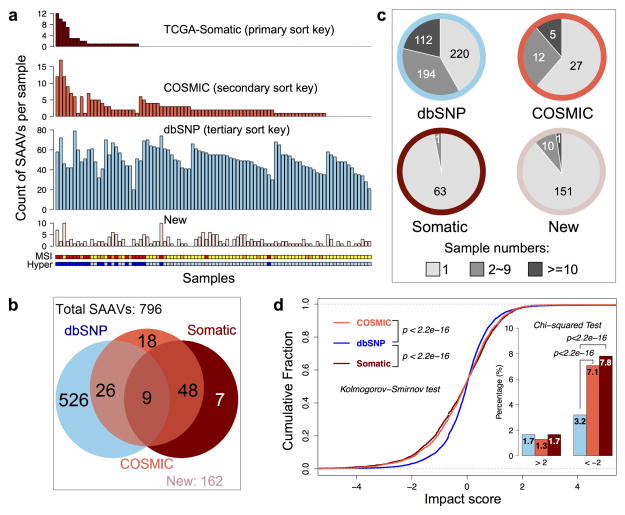
Figure 1. Summary of detected single amino acid variants (SAAVs) and the impact of single nucleotide variants (SNVs) on protein abundance.
a, The number of different types of SAAVs (TCGA-reported somatic variants, COSMIC-supported variants, dbSNP-supported variants, and new variants) in individual tumor samples. The samples are ordered by the number of detected somatic variants, then COSMIC-supported variants, and then dbSNP-supported variants. The Microsatellite instability (MSI) and hypermutation (Hyper) status are labeled below the bar charts for each sample (MSI-High: red, MSI-Low: orange, Microsatellite Stable: yellow; hypermutated: blue, non-hypermutated: sky blue; no data: grey). The number of somatic variants and COSMIC-supported variants were significantly higher in MSI-High and hypermutated tumors, whereas the other two types of SAAVs were randomly distributed across the data set. b, The total numbers for different types of SAAVs and their overlapping relations. All 796 detected SAAVs were annotated based on previous reports in dbSNP (left circle), COSMIC (middle circle), or TCGA-reported somatic variants (right circle), and their overlapping relations are shown in the Venn diagram. There are 162 SAAVs that have not been reported previously in these databases (new). c, Distribution of the frequency of occurrence (1 sample: light grey, 2–9 samples: grey, >=10 samples: dark grey) for different types of SAAVs. Border colors of the pie charts correspond to different SAAV types using the same color scheme as in (a). Whereas 58% of dbSNP-supported variants occurred in two or more samples, almost all somatic variants each occurred in only one sample. d, SNVs detected in RNA-Seq data were separated into three categories (dbSNP-supported, COSMIC-supported, and TCGA-Somatic). The impact of individual SNVs on protein abundance was calculated (see supplementary methods) and the impact scores for different categories of SNVs were plotted as cumulative fraction curves with two-sided p values from the Kolmogorov-Smirnov test labeled. The percentage of SNVs with an absolute impact score greater than 2 was also plotted as an inset, with p values from the Chi-squared test. Sample size for the dbSNP-supported, COSMIC-supported and TCGA-Somatic variants were 12184, 7492, and 3302, respectively.