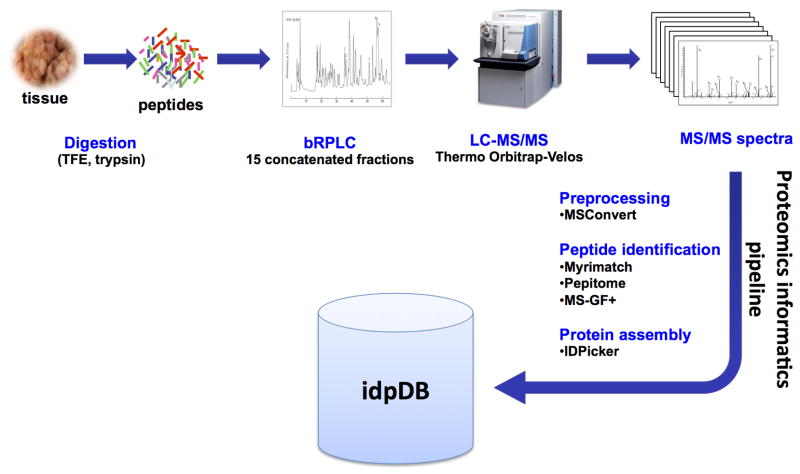
Extended Data Figure 1. Mass spectrometry (MS)-based proteomics workflow.
Protein was extracted from frozen tumor tissue and used to generate tryptic digests. The resulting tryptic peptides were fractionated using off-line basic-reverse phase high-pressure liquid chromatography (bRPLC). Collected fractions were pooled and used for reverse phase HPLC in-line with a Thermo Orbitrap-Velos MS instrument. Raw data was processed by MSConvert and then used for database and spectral library searching using three different search engines (Myrimatch, Pepitome and MS-GF+). Identified peptides were assembled using IDPicker 3 with selected filters as described in the methods. IDPicker 3 stores its protein assemblies for a specified set of filters in the idpDB format. These SQLite databases associate spectra with peptides, peptides with proteins, and LC-MS/MS experiments with a hierarchy of experiments.