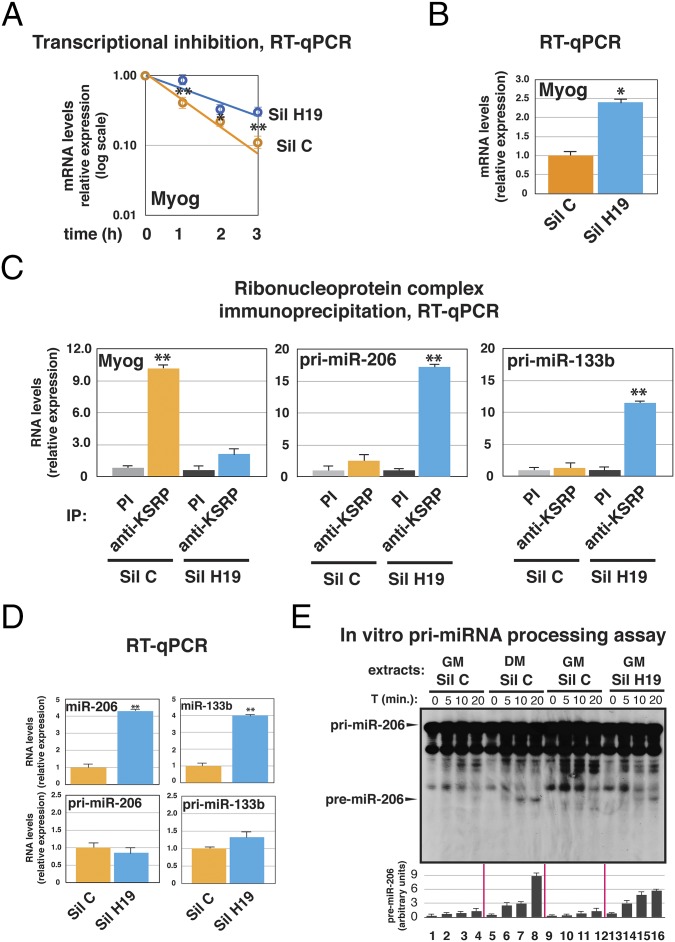
Fig. 3.
H19 silencing in undifferentiated C2C12 cells promotes myogenin mRNA stabilization and maturation of myomiRs from precursors. H19 was transiently silenced in C2C12 cells cultured in GM by using a combination of a sequence-specific siRNA and a GAPmeR (collectively indicated as Sil H19). Parallel cultures were mock-silenced by using a combination of a control siRNA and a control GAPmeR (collectively indicated as Sil C). Cells were used for experiments 48 h after transfection. (A) Cells were treated with 100 μM 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB), and total RNA was isolated at different times (as indicated) after the addition of DRB and analyzed by RT-qPCR to detect myogenin mRNA expression. (B) RNA was prepared from transfected C2C12 cells, and myogenin levels were quantified by RT-qPCR. (C) Total cell extracts were immunoprecipitated as indicated. RNA was purified from immunocomplexes and analyzed by RT-qPCR to detect myogenin or primary miRNAs. (D) RNA was prepared from transfected C2C12 cells, and the levels of the indicated miRNAs and primary miRNAs were quantified by RT-qPCR. (E) In vitro pri-miR-206 processing assays performed by using total extracts from C2C12 cells mock- (Sil C) or H19- (Sil H19) silenced and cultured in GM or DM (for 36 h). Internally 32P-labeled pri-miR-206 RNA substrate was added, and its processing was monitored as described under Materials and Methods. A representative autoradiogram is shown. The intensity of background bands is ascribable to the long exposure of gels as a result of the low processing efficiency of pri-miR-206. The arrow points to premiR-206 band that is visible only in experiments performed by using extracts from cells treated with DM/Sil C or GM/Sil H19. The bar graph below the autoradiogram is a quantification of the premiR-206 levels measured in two distinct processing assays. The intensity of the bands corresponding to pri–miR-206 and premiR-206, quantified with ImageJ software (http://rsb.info.nih.gov/ij/index.html), was expressed as percentage (±SEM calculated on two experiments) of premiR-206 generated from pri-miR-206 at each time point. To avoid signal saturation, the quantification was performed on underexposed autoradiograms. The values of RT-qPCR experiments shown are averages (±SEM) of three independent experiments performed in triplicate (*P < 0.01 and **P < 0.001, Student t test).