Figure 4.
The Loss of PRMT5 in PGCs Induces p53 Signaling and DNA Damage Response Genes
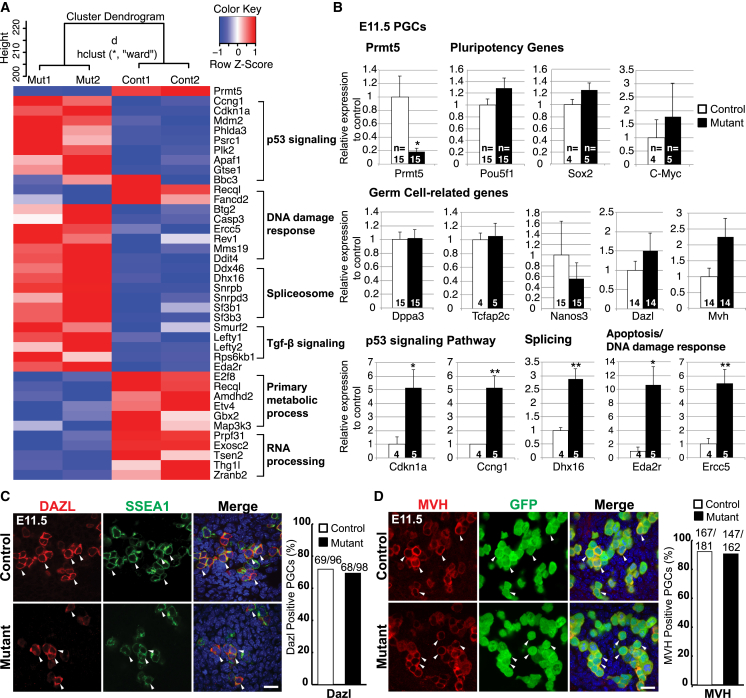
(A) A heatmap of selected up- and downregulated genes in E11.5 PGCs from RNA-Seq analysis. Cluster dendogram for two biological replicates of each genotype is shown on top. Note that p53 signaling genes and DNA damage response genes are upregulated (red) in the mutant, while primary metabolic process and RNA processing genes are downregulated (blue).
(B) qRT-PCR analysis of selected genes in E11.5 PGCs (control, white; mutant, black). Shown are the mean values ± SE. Significance is shown by Student’s t test: ∗∗p < 0.01, ∗p < 0.05.
(C) IF staining of DAZL (late germ cell marker, red) in genital ridges at E11.5. PGCs were detected with antibodies against SSEA1 (green). Merged images are shown with DNA stained with DAPI (blue). The ratio of DAZL-positive PGCs in SSEA1-positive PGCs (in %) was determined (controls, white; mutants, black).
(D) IF staining of MVH (late germ cell marker, red) in genital ridges at E11.5. The ratio of MVH-positive PGCs in GFP positive (green) PGCs (in %) was determined. Scale bar, 20 μm in (C) and (D). The arrowheads in (C) and (D) indicate examples of PGCs. The genotype of the control is Blimp1Cre;Prmt5flox/+, and the mutant is Blimp1Cre;Prmt5flox/− in (A)–(D).