Abstract
Cell signaling underlies critical cellular decisions. Coordination, efficiency as well as fail-safe mechanisms are key elements. How the cell ensures that these hallmarks are at play are important questions. Cell signaling is often viewed as taking place through discrete and crosstalking pathways; oftentimes these are modularized to emphasize distinct functions. While simple, convenient and clear, such models largely neglect the spatial structure of cell signaling; they also convey inter-modular (or inter-protein) spatial separation that may not exist. Here our thesis is that cell signaling is shaped by a network of multiprotein assemblies. While pre-organized, the assemblies and network are loose and dynamic. They contain transiently-associated multiprotein complexes which are often mediated by scaffolding proteins. They are also typically anchored in the membrane, and their continuum may span the cell. IQGAP1 scaffolding protein which binds proteins including Raf, calmodulin, Mek, Erk, actin, and tens more, with actin shaping B-cell (and likely other) membrane-anchored nanoclusters and allosterically polymerizing in dynamic cytoskeleton formation, and Raf anchoring in the membrane along with Ras, provides a striking example. The multivalent network of dynamic proteins and lipids, with specific interactions forming and breaking, can be viewed as endowing gel-like properties. Collectively, this reasons that efficient, productive and reliable cell signaling takes place primarily through transient, preorganized and cooperative protein-protein interactions spanning the cell rather than stochastic, diffusion-controlled processes.
Keywords: cell organization, cell signaling, signaling pathways, signal transduction, cell structure, signaling modules, diffusion
1. Introduction
A living cell is an organized pattern, structured in space and time (Bolanos-Garcia et al., 2012; Harold, 2005; Nussinov, 2013). Architecture is what ultimately distinguishes a living cell from some haphazard assemblage in solution (McLaughlin et al., 2012). How a cell achieves, preserves, and replicates its spatial organization and how dynamic viable signaling persists within it are fundamental to the understanding of the living state. The cellular architecture is important for the cell’s mechanical properties, morphology, motility, metabolism, supramolecular order, chromatin organization and gene expression, trafficking and more. It is also crucial for signaling within and between cells. Signals propagate through interactions; chief among these are between proteins. The cellular organization is hierarchical. Notwithstanding, there is a continuum from small molecular complexes to nanoclusters and membrane domains, to the cytoskeleton (Chen et al., 2014a; Chia et al., 2014); from cell-to-cell interface, to the membrane to the cytoplasm and to the organelles. Such multi-scale organization feeds back to regulate specific proteins, and collectively cell signaling; and at the basic level it does so through dynamic reorganization of multiprotein complexes and assemblies. Dynamic multiprotein complexes are the fundamental unit of cellular organization and signaling. Transient complexes hold the key for the ability of the cell to survive and to respond to its changing environment. Dynamic association implies not merely interactions forming and dissociating; it connotes cooperativity which can specify which interaction takes place at any given time at a given shared binding site. Within this framework, cell signaling can be viewed in terms of dynamic allosteric interactions within and among, spatially organized transient multimolecular complexes. The complexes vary over time and space. A key challenge is to understand the interplay across these complexes, link it to the physicochemical basis of the conformational behavior of single molecules, and ultimately relate it to global cellular function. Overall, our thesis is that cell signaling should be thought of as transient, allostery-driven forming and reforming interactions taking place within dynamic, loosely preorganized assemblies, rather than as a sequence of diffusion-controlled molecular collisions (Nussinov, 2013). Growth, differentiation, division, and apoptosis, are temporal; they can be understood only in terms of dynamics within, and among, assemblies and multiprotein complexes. And within this framework, coordination is governed by a conformational biasing mechanism, that is, population shift (Dixit and Verkhivker, 2011; Gunasekaran et al., 2004; Kar et al., 2010; Kumar et al., 2000; Long and Bruschweiler, 2011; Ma et al., 1999; Ma et al., 2002; Rivalta et al., 2012; Shan et al., 2012; Tsai et al., 1999a; Tsai et al., 1999b; Tsai et al., 2001). Population shift is the origin of allostery; it is the means through which action at the surface of one protein can be expressed by another, far away.
Proteins are often viewed as freely diffusing in the cell. This leads to questions such as how molecules efficiently find their proper location in cell space (Nussinov, 2012). In contrast, here we view cellular signaling as transient pre-organized and inter-connected protein assemblies which span the cell, with signaling taking place via dynamic conformational population shifts (Bolanos-Garcia et al., 2012; Nussinov, 2013). Such a multivalent, typically membrane-anchored network, with interactions forming and breaking endows cell signaling with gel-like properties (Figure 1). We reason that this may well be the efficient, robust, cooperative and controlled signaling system embraced by evolution.
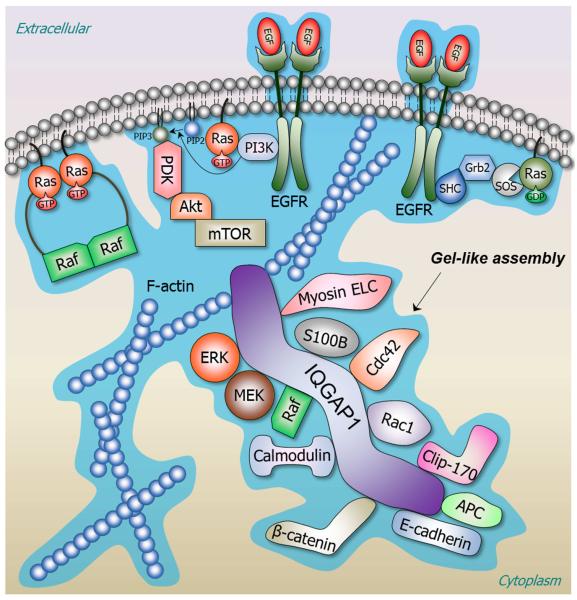
Figure 1.
Cellular signaling through pre-organized and inter-connected nanocluster assemblies with gel-like properties. Examples of clusters illustrate Ras activation and scaffolding proteins, such as IQ motif containing GTPase activating protein 1 (IQGAP1). Ras is a small GTPase that is related to numerous cellular functions, including cell proliferation, apoptosis, migration, fate specification, and differentiation. A key Ras effector pathway is the mitogen-activated protein kinase (MAPK) pathway, i.e. Raf/MAPK kinase/extracellular signal-regulated kinase (Raf/MEK/ERK) pathway. Ras is normally activated in response to the binding of extracellular ligands, such as epidermal growth factor (EGF), to a receptor tyrosine kinase (RTK), e.g. epidermal growth factor receptor (EGFR). The signal triggered by EGF binding to the extracellular domain of EGFR is transmitted through the transmembrane domain resulting in EGFR dimerization and activation. Signalling proceeds through SHC-transforming protein 1 (SHC), growth factor receptor-bound protein 2 (Grb2) and son of sevenless (SOS), the Ras guanine nucleotide exchange factor (GEF). GEF exchanges GDP by GTP, activating Ras. Active, GTP-loaded Ras dimerizes and binds Raf, thereby promoting Raf dimerization and activation. Active Raf dimer phosphorylates and activates MEK, which induces ERK activation. Ras activates a number of signalling pathways. Ras in the active GTP-bound state regulates phosphatidylinositol 3-kinase (PI3K) pathway. PI3K is a heterodimer containing a regulatory (p85) and catalytic (p110) subunits. RTKs recruit the p85 subunit of PI3K. Ras activates p110 independently of p85. PI3K phosphorylates phosphatidylinositol-4,5-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-trisphosphate (PIP3), which recruits phosphoinositide-dependent kinase-1 (PDK1) and phosphorylates a serine/threonine kinase, Akt (also known as PKB, protein kinase B) in the plasma membrane. This further induces the activation of mammalian target of rapamycin (mTOR) complex, one of the major pathways leading to cell growth. A scaffolding protein, IQGAP1, a large multi-domain protein binds to tens of partners and regulates the function of numerous proteins. Over 90 proteins have been reported to associate with IQGAP1, directly or as part of a larger complex. These include cytoskeleton proteins actin and myosin, as well as other proteins, which are also partners of IQGAP1. Actin is a scaffolding protein too with many partners and also anchors to the membrane such as Ras and Raf. As figure shows, IQGAP1 is a scaffold in the MEK/ERK cascade and binds to many proteins including Raf, calmodulin, and S100 calcium binding protein B (S100). Multivalent membrane-anchored proteins and intracellular multiproteins form gel-like complexes in a network with transient connections. Abbreviations used in the figure are: APC, adenomatous polyposis coli; Cdc45, cell division control protein 42 homolog; Clip-170, cytoplasmic linker protein-170; Rac1, Ras-related C3 botulinum toxin substrate 1.
2. A view of cellular organization
Cells are often considered and drawn schematically with proteins encased in a modular organization (Chen et al., 2014b; Resendis-Antonio et al., 2012; Roy et al., 2013; Sohn et al., 2011; Song and Singh, 2013). The underlying premise is that within modules the proteins are likely to be in spatial vicinity, unlike between modules. However to function the module composition needs to change dynamically. Proteins from one module would also need to interact- directly or indirectly- with those in another. Function builds on signaling within and between modules, which can only take place via physical interactions. Here we contend that evolution is unlikely to have cellular communication rely on random diffusion across the large distances in the cell. A random process can place in during basal expression or module (or cluster) re-association of proteins nearby; it is not likely to be productive if the modules are far away. As a cellular signaling mechanism, long distance diffusion can become even more questionable when we consider that cells are organized and structured. They consist of membrane-enveloped organelles and cytoplasm; with functional units either attached to the membrane or partitioned and localized by cytoskeleton proteins (Chen et al., 2014a; Chia et al., 2014). Such a high level of organization optimized by evolution does not appear compatible with micrometer scale diffusion-controlled signaling. Current data point to signaling pathways as complex sets of ordered events; stochastic long distance diffusion would dampen cellular response. The volume excluded by the cytoskeleton increases the crowding and thus the intermolecular association constants which may suggest the feasibility of interaction during a ‘random walk’ in open space. Such random walks are concentration-dependent, and rely on the ability to move rapidly over long distances (Cebecauer et al., 2010). In contrast, the subnanomolar concentration of growth factors triggering cell stimulation and the concentration of membrane-bound ligands that provoke cellular responses suggest that signaling molecules interact at low concentrations.
Cellular processes need to be regulated. Regulation requires efficiency. Here, we distinguish between multimolecular assemblies and complexes. We define assemblies as large, loose and dynamic multimolecular associations. The assemblies are transient, and freely diffusing molecules may shift to form new assemblies. The molecules are in binding-competent states, poised for direct productive interaction. The assemblies embody smaller multimolecular complexes, which we define as physically interacting molecules. The complexes are similarly transient, dissociating and re-associating, responding to and transmitting cell signals. Clusters can be described as larger and looser bodies such as those of proteins anchored in the membrane rafts and containing lipids. Ras nanoclusters provide one example (Harding and Hancock, 2008; Janosi et al., 2012); T- and B-cell receptors provide another (Molnar et al., 2012). All promote heterogeneous molecular landscapes. Signaling proceeds through a population shift mechanism of the proteins across dynamically pre-organized assemblies, via the direct physical interfaces of the complexes, or mediated by other molecular types, including lipids and water.
While both the population shift and the diffusion-controlled chance collisions mechanisms can co-exist and are not mutually exclusive, signaling is likely to be more productive in pre-organized states (Nussinov, 2013). Accompanied by factors such as protein concentration, cofactors and metabolites, and membrane composition, these may offer an explanation how despite cellular complexity, the cell accomplishes coordination and potent response. The merit of such a view is that it underscores dynamic, transient associations of multiprotein complexes, and provides for a continuum in cell space via inter-connected assemblies and clusters. As we discuss below, these largely take place via scaffolding proteins which bind a large number of partners with different functions; and they do so across a range of scales, including the cytoskeleton (Head et al., 2014). This is amplified by interactions between their partners which are often further supplemented and mediated by the plasma (or organelle) membranes, nucleic acids, ions, water and small molecules such as metabolites and hormones.
3. Multiprotein complexes and the role of allostery
Multiprotein complexes are common in the cell. They fulfill a broad range of functions. They typically contain several enzymes catalyzing successive reactions and in higher organisms are often mediated by scaffolding (or adaptor) proteins as well. The MAPK complex and the E3 system in ubiquitination are two examples. As the case of the KSR1 shows, scaffolding proteins may also function as enzymes (Zhang et al., 2013). Allostery plays a key role in the presence and in the absence of scaffolding proteins (Nussinov et al., 2013a). In the absence of scaffolding proteins, the precise, often short-lived, topographical organization of the enzymes in the complex allows allosteric propagation through the enzyme-enzyme interface which can prime successive enzymatic reactions. A precise organization in the complex is critical; a mere co-localization of the enzymes is non-specific, and cannot achieve such coordination. Nonetheless, such an enzyme-only organization is limited in the range of cellular functional coordination that it can achieve. That however is not the case if scaffolding proteins exist in the complex. Scaffolding proteins can link functions, regulate pathway cross-talk and allow more complex cellular control. Metabolic multiprotein complexes do not appear to contain scaffolding proteins; however, signaling multiprotein complexes usually do. Scaffolding proteins are essential for signal transfer and manipulation. They are active components of multienzyme complexes; much more so than considered by the classical view. Traditionally scaffolding proteins were postulated to act as a co-localizing platform for their partners. We suggested that the platform is dynamic, and decides pathway activation or inhibition. Since a scaffolding protein typically binds a large number of partners, which can be in the tens or in the hundreds, binding sites are shared. Partner selection is decided by three factors: effective local concentration, distinct combination of post-translational modifications (PTMs) at the binding site (if any), and prior allosteric effects, either induced by allosteric PTMs (Nussinov et al., 2012) or by earlier binding events. Collectively, these tag the binding site with a specific barcode which serves as an address label (Nussinov et al., 2013b; Nussinov et al., 2013c).
Allostery plays a key role in dynamically and efficiently controlling signals (Ma and Nussinov, 2009; Nussinov, 2012). It does so via population shift (Kar et al., 2010; Liu and Nussinov, 2011; Pandini et al., 2012). Allostery is selective: the perturbations elicited by specific effectors lead to increased (or, decreased) populations of distinct conformational states whose active sites favor (disfavor) certain partners (Cui and Karplus, 2008; Laine et al., 2012; Lechtenberg et al., 2012; Ma et al., 2010). It can be reflected by changes in conformation or in dynamics (del Sol et al., 2009; Kenakin and Miller, 2010; Tsai et al., 2008; Tsai et al., 2009; Tzeng and Kalodimos, 2011; Zhuravlev and Papoian, 2010). Multiprotein complexes form a network with transient connections (Figure 1) that control diverse cellular processes. Below, we detail some examples of multiprotein complexes that play key roles in human signaling pathways. They contain enzymes and scaffolding proteins. Within this framework, we highlight the role of allostery in favoring certain interactions over others, and by so doing decide which pathway to follow under certain conditions. Allostery is a fundamental physical phenomenon in the cell; however, it is challenging to predict its consequences. This is particularly the case in real scenarios, where at any given time multiple allosteric events take place in a multiprotein complex.
4. Multiprotein complexes may be anchored in membrane domains
While here we focus on protein-protein interactions, membrane domains can play a fundamental role in their types, composition and organization. Multiprotein complexes can either anchor directly in the membrane, or indirectly through loosely and dynamically interconnected multiprotein assemblies. The membrane composition is not homogeneous across local membrane rafts, resulting in varied protein assemblies. A case in point is the Ras isoforms: K-, N, and HRas vary in their preferences, leading to segregation of their nanoclusters. Of interest, Raf preferentially anchors and spends more time in disordered negatively charged domains, which are those preferred by the mutationally highly oncogenic K-Ras isoform (Cho et al., 2012; Janosi et al., 2012). On a different note, nucleic acids also play key roles in anchoring multiprotein complexes. Chromatin, transcription initiation complexes, and the large ribosomal translational machineries can provide some examples.
5. 14-3-3 and IQGAP1 scaffolding proteins examples
14-3-3 provides one example of a scaffolding protein which bridges a large number of transient multi-protein complexes in dynamic assemblies; the large, multi-domain IQGAP1 which participates in multiple essential functions of mammalian biology by binding to and regulating numerous interacting proteins is another (White et al., 2012). Over 90 proteins have been reported to interact with IQGAP1 either directly or indirectly. All likely participate in its transient multiprotein complexes. Its interactions mediate receptors and their signaling cascades, small GTPases, cytoskeletal dynamics, neuronal regulation and intracellular trafficking (Figure 1). 14-3-3s are phospho-serine/threonine binding proteins with the ability to bind multiple, functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors. Unlike IQGAP1, they are small acidic proteins with a molecular mass ranging from 27 to 32 kDa. Typical of scaffolding proteins, they have no detectable catalytic domain or function. More than 200 signaling proteins are currently suggested to be 14-3-3 ligands, although the actual number of confirmed ligands is around 70. Among these, recent analysis indicated that 14-3-3s dock onto 40% of human kinases, many more than has been realized (Tinti et al., 2014). They are dimeric proteins with two antiparallel ligand binding grooves. The dimeric structure of the 14-3-3 protein allows it to bind two ligands simultaneously. The dimer is arranged such that the ligand binding groove runs in opposite directions in each monomer of the molecule. In the co-crystal structures, the ligand binding sites are located within the same concave surface, and each site is occupied. Many binding partners contain two or more 14-3-3 binding motifs, which can bind simultaneously to both ligand grooves (Johnson et al., 2010; Kostelecky et al., 2009). Common determinants among the ligands are phosphorylated motifs. As typical for phosphorylation sites, these regions are disordered (Bozoky et al., 2013; Tyanova et al., 2013). Like all adapter proteins, the 14-3-3 induce conformational change in their ligands. What regulates the binding of 14-3-3 to any specific ligand at any specific time is still unclear. We may assume that at least two factors are involved: the presence of their ligand proteins in the phosphorylated state in their multiprotein assembly, poised for binding, and prior interaction and (or) post-translational modifications (PTMs), priming its binding site for a specific binding-ready conformation complementary to a distinct ligand. Even though the structure of the 14-3-3 has been postulated to be rigid, like all proteins, 14-3-3 can be expected to exist as conformational ensemble of states, with some distributions. Following binding or allosteric PTM events, population shift will take place, making it binding-competent. 14-3-3 ligands (Tzivion et al., 2001) include the PKCε (the V3 region contains two adjacent phosphorylated 14-3-3 binding motifs), proteins involved in cell cycle control such as Cdc25 (Conklin et al., 1995; Peng et al., 1997), Wee1 (Honda et al., 1997), p53 (Waterman et al., 1998), CDC2 (Chan et al., 1999), CDK2 (Laronga et al., 2000) and the centrosome structure (Pietromonaco et al., 1996), proteins involved in cellular signaling and stress response like Raf (Fantl et al., 1994; Freed et al., 1994; Fu et al., 1994; Irie et al., 1994), IGF-I receptor (Craparo et al., 1997; Furlanetto et al., 1997), IRS-1 (Craparo et al., 1997; Ogihara et al., 1997), PI-3 kinase (Bonnefoy-Berard et al., 1995), PKC (Aitken et al., 1995), Cbl (Liu et al., 1996), Bcr (Reuther et al., 1994), polyoma middle T antigen (Pallas et al., 1994), MEKK-1 and 4 (Fanger et al., 1998), MLK2 (Nagata et al., 1998), BAD (Xiao et al., 1995) and ASK-1 (Xiao et al., 1995), transcription regulation such as FKHRL1 (Brunet et al., 1999), DAF-16 (Cahill et al., 2001), TAZ (Kanai et al., 2000), TLX-2 (Tang et al., 1998) and histone deacetylase (Grozinger and Schreiber, 2000; Wang et al., 2000) and cytoskeletal proteins like keratin K18 (Liao and Omary, 1996) and vimentin (Tzivion et al., 2000). Regulation of binding can be seen in the Raf-1 example (Fu et al., 2000). The prototype phosphorylated serine recognition motif, RSxpSxP. In 14-3-3 it is RQRS257TS259TP. When phosphorylated on S259, it binds 14-3-3f with an apparent Kd of 122 nM. However, when unphosphorylated or when phosphorylated at S257 or at both S257 and S259 it cannot bind. A-, B-, and C-RAF activity is differentially regulated by its C-terminal and internal 14-3-3 binding domain (Fischer et al., 2009). Serine residues are at positions 43, 259, and 621. 259 and 621 are involved in binding of 14-3-3 proteins to C-RAF. Phosphorylation of serine 621 appears essential for C-Raf activation. Phosphorylation of serine 259 is inhibitory (Freed et al., 1994; Zhang et al., 1999).
14-3-3 plays a critical regulatory role in the Ras-Raf signaling pathway. It’s binding to the inactive closed Raf conformation triggers an allosteric conformational change, which permits the binding of Raf’s Ras binding domain (RBD) to Ras. In turn, this results in Raf’s allosterically modulating the conformation of its catalytic domain, priming it to dimerization, and Raf’s activation. Raf’s cysteine-rich domain (CRD) is also believed to bind Ras, a G protein, as well as the membrane. Raf-CRD can bind 14-3-3 in addition to Ras and pS259. In contrast to the pS259 and CRD sites, the interaction of 14-3-3 with Raf-1 at the pS621 site may be required for Raf-1 activation. Fischer et al. (Fischer et al., 2009) suggested a model where that association of C-Raf with Ras-GTP and membrane lipids results in translocation of inactive cytosolic doubly-bound (at 259 and 621) RAF/14-3-3 complex to the plasma membrane. Subsequent interaction of membrane-bound prohibitin (PHB) with C-RAF near Raf’s isoform-specific hinge segment region (Baljuls et al., 2008) displace 14-3-3 from the internal binding domain at Ser 259, allowing access to phosphatases with subsequent dephosphorylation. Prohibitin is a scaffolding protein that interferes with the internal Ser259 14-3-3 binding site in C-Raf. Interestingly, the crystal structure of a phosphorylated, farnesylated peptide from Rnd3, another G-protein, with 14-3-3 revealed that the hydrophobic groove in 14-3-3 proteins accommodates the farnesyl moiety along with the phosphorylated motif (Riou et al., 2013).
6. Multiprotein assemblies shape the spatial structure of cell signaling
Altogether, a complex picture emerges of protein assembly in the cell. It emphasizes also the distinction between protein complexes and assemblies in the cell. Direct complexes take place between 14-3-3 and Raf. At the same time 14-3-3 is also a dimer. Prohibitin appears transiently bound as well in the case of C-Raf. Other isoforms may have other scaffolding proteins fulfilling such roles. Raf also interacts with Ras, possibly through two of its domains. Raf is also anchored in the membrane as is Ras. Ras also forms nanoclusters, through its membrane-anchored farnesylated C-terminal hypervariable regions. Protein kinase C (PKC), also a ligand of 14-3-3, is responsible for phophorylating K-Ras4B at Ser181. Phosphorylation is required for tumor growth (Barcelo et al., 2014). Dephosphorylation by a phosphatase (e.g. λ phosphatase) results in calmodulin binding and inhibition of this Ras-Raf pathway. Direct interaction between calmodulin and Ras has been detected (Abraham et al., 2009; Alvarez-Moya et al., 2011; Chavan et al., 2013). Further, Raf and calmodulin are known to directly bind to IQGAP1 as are Erk and Mek, cadherin, actin and APC (Malarkannan et al., 2012). Erk and Mek are key kinases in the EGFR-Ras-Raf-Mek-Erk MAPK pathway. Caveolin-1 and IQGAP1 scaffolding proteins are required for phosphorylation of the actin associated pool of ERK1/2 in response to protein kinase C activation (Vetterkind et al., 2013). Cadherins are calcium-dependent adhesion transmembrane proteins; actin is an allosteric dynamic protein which forms microfilaments, dynamically polymerizes and depolymerizes, and constitutes a key component of the cytoskeleton. Actin is also established to play a key role in formation of B-cell receptors nanoclustering. Together with tetraspanin transmembrane scaffolding proteins it forms networks that organize receptor nanoclusters to regulate B cell receptor-mediated signaling (Mattila et al., 2013). APC plays a critical role in several cellular processes that determine whether a cell may develop into a tumor (Minde et al., 2011) controlling how often a cell divides, how it attaches to other cells within a tissue, or whether a cell moves within or away from a tissue. In particular, β-catenin in the Wnt signaling pathway is controlled by the APC protein.
7. Conclusions and outlook
Over the last few years, considerable literature addressed the hallmarks of protein-protein interactions, along with emerging strategies for their inhibition. An increasing number of publications have also focused on constructing their networks in the cell and their connectivity (Hou et al., 2014). These embraced regulatory networks (Chetverina et al., 2014), networks of diseases (Elias et al., 2014; Soldevilla et al., 2013), protein interaction networks in immunity (Androulakis et al., 2013; Nibbs and Graham, 2013), ErbB network (Sanchez-Soria and Camenisch, 2010; Seshacharyulu et al., 2012), cell surface’s protein network (Welz et al., 2014), NF-κB signaling network (Konrath et al., 2014), networks of polypharmacology (Engin et al., 2014), network characteristics that correlate environmental and genetic robustness (Shreif and Periwal, 2014), personalized mutation network (Jia and Zhao, 2014) and more. Some are represented by node-and-edge graphs; others are mode detailed, structural networks (Acuner Ozbabacan et al., 2014; Kuzu et al., 2013). Together, these present a comprehensive and ever growing coverage.
Here we sought to project protein-protein interactions in the framework of the cell. Neither protein-protein interactions nor their network representations capture the cell’s organization. Networks are depicted as cell-spanning, largely overlooking the fact that proteins typically function not in binary interactions but while associated as multimolecular complexes; when confined in some modular representation, the modules are distinct and separated. Neither representation fully grasps the continuum in the cell. This is important, since drawings may influence a reader’s perception.
Here we suggested that cell is pre-organized. Pre-organization does not imply an immobile behavior; the distinct intermolecular interactions fluctuate with short residence timescales. The basic units consist of transient multiprotein complexes. These short-lived interactions allow coordination and priming successive enzymes in catalytic pathways. Scaffolding proteins play major roles. They not only communicate the signals, but can control it. The large number of possible partners that they, and other hub proteins, bind argues that the complexes can be viewed as residing within larger, loose, interconnected and highly dynamic assemblies. The IQGAP1 scaffolding described above, which links membrane-anchored proteins in key signaling pathways with allosteric filamentous cytoskeleton proteins which at the same time act to regulate and organize nanoclusters in the membrane provides a striking example of how dynamic networked protein-protein interactions can shape cell signaling systems (Figure 1). Such a pre-organized, yet dynamic view of signaling in the cell underscores efficiency. It reasons that productive interactions are unlikely to take place via chance collisions. A population shift mechanism among pre-organized, albeit highly mobile molecules in an environment endowed by gel-like properties, appears a more plausible strategy to have been adopted by evolution. Along these lines, data have been suggested in favor caveolin-1 and IQGAP1 assembling distinct signaling complexes, which are then dynamically linked in a hierarchical arrangement to generate a functional ERK1/2 activation pathway (Vetterkind et al., 2013).
Acknowledgements
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
9. References
- Abraham SJ, Nolet RP, Calvert RJ, Anderson LM, Gaponenko V. The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry. 2009;48:7575–83. doi: 10.1021/bi900769j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuner Ozbabacan SE, Gursoy A, Nussinov R, Keskin O. The structural pathway of interleukin 1 (IL-1) initiated signaling reveals mechanisms of oncogenic mutations and SNPs in inflammation and cancer. PLoS computational biology. 2014;10:e1003470. doi: 10.1371/journal.pcbi.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken A, Howell S, Jones D, Madrazo J, Martin H, Patel Y, Robinson K. Post-translationally modified 14-3-3 isoforms and inhibition of protein kinase C. Molecular and cellular biochemistry. 1995;149-150:41–9. doi: 10.1007/BF01076562. [DOI] [PubMed] [Google Scholar]
- Alvarez-Moya B, Barcelo C, Tebar F, Jaumot M, Agell N. CaM interaction and Ser181 phosphorylation as new K-Ras signaling modulators. Small GTPases. 2011;2:99–103. doi: 10.4161/sgtp.2.2.15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androulakis IP, Kamisoglu K, Mattick JS. Topology and dynamics of signaling networks: in search of transcriptional control of the inflammatory response. Annual review of biomedical engineering. 2013;15:1–28. doi: 10.1146/annurev-bioeng-071812-152425. [DOI] [PubMed] [Google Scholar]
- Baljuls A, Schmitz W, Mueller T, Zahedi RP, Sickmann A, Hekman M, Rapp UR. Positive regulation of A-RAF by phosphorylation of isoform-specific hinge segment and identification of novel phosphorylation sites. The Journal of biological chemistry. 2008;283:27239–54. doi: 10.1074/jbc.M801782200. [DOI] [PubMed] [Google Scholar]
- Barcelo C, Paco N, Morell M, Alvarez-Moya B, Bota-Rabassedas N, Jaumot M, Vilardell F, Capella G, Agell N. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer research. 2014;74:1190–9. doi: 10.1158/0008-5472.CAN-13-1750. [DOI] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Wu Q, Ochi T, Chirgadze DY, Sibanda BL, Blundell TL. Spatial and temporal organization of multi-protein assemblies: achieving sensitive control in information-rich cell-regulatory systems. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences. 2012;370:3023–39. doi: 10.1098/rsta.2011.0268. [DOI] [PubMed] [Google Scholar]
- Bonnefoy-Berard N, Liu YC, von Willebrand M, Sung A, Elly C, Mustelin T, Yoshida H, Ishizaka K, Altman A. Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3-3 proteins in T cells. Proc Natl Acad Sci U S A. 1995;92:10142–6. doi: 10.1073/pnas.92.22.10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozoky Z, Krzeminski M, Chong PA, Forman-Kay JD. Structural changes of CFTR R region upon phosphorylation: a plastic platform for intramolecular and intermolecular interactions. The FEBS journal. 2013;280:4407–16. doi: 10.1111/febs.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. The Journal of biological chemistry. 2001;276:13402–10. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- Cebecauer M, Spitaler M, Serge A, Magee AI. Signalling complexes and clusters: functional advantages and methodological hurdles. Journal of cell science. 2010;123:309–20. doi: 10.1242/jcs.061739. [DOI] [PubMed] [Google Scholar]
- Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–20. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- Chavan TS, Abraham S, Gaponenko V. Application of reductive (1)(3)C-methylation of lysines to enhance the sensitivity of conventional NMR methods. Molecules. 2013;18:7103–19. doi: 10.3390/molecules18067103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014a;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Z, Wang Y. Spatiotemporal positioning of multipotent modules in diverse biological networks. Cellular and molecular life sciences : CMLS. 2014b doi: 10.1007/s00018-013-1547-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays : news and reviews in molecular, cellular and developmental biology. 2014;36:163–72. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PH, Chen B, Li P, Rosen MK, Shen K. Local F-actin network links synapse formation and axon branching. Cell. 2014;156:208–20. doi: 10.1016/j.cell.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KJ, Kasai RS, Park JH, Chigurupati S, Heidorn SJ, van der Hoeven D, Plowman SJ, Kusumi A, Marais R, Hancock JF. Raf inhibitors target ras spatiotemporal dynamics. Current biology : CB. 2012;22:945–55. doi: 10.1016/j.cub.2012.03.067. [DOI] [PubMed] [Google Scholar]
- Conklin DS, Galaktionov K, Beach D. 14-3-3 proteins associate with cdc25 phosphatases. Proc Natl Acad Sci U S A. 1995;92:7892–6. doi: 10.1073/pnas.92.17.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craparo A, Freund R, Gustafson TA. 14-3-3 (epsilon) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. The Journal of biological chemistry. 1997;272:11663–9. doi: 10.1074/jbc.272.17.11663. [DOI] [PubMed] [Google Scholar]
- Cui Q, Karplus M. Allostery and cooperativity revisited. Protein science : a publication of the Protein Society. 2008;17:1295–307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Sol A, Tsai CJ, Ma B, Nussinov R. The origin of allosteric functional modulation: multiple pre-existing pathways. Structure. 2009;17:1042–50. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Verkhivker GM. Computational modeling of allosteric communication reveals organizing principles of mutation-induced signaling in ABL and EGFR kinases. PLoS computational biology. 2011;7:e1002179. doi: 10.1371/journal.pcbi.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J, Dimitrio L, Clairambault J, Natalini R. The p53 protein and its molecular network: modelling a missing link between DNA damage and cell fate. Biochimica et biophysica acta. 2014;1844:232–47. doi: 10.1016/j.bbapap.2013.09.019. [DOI] [PubMed] [Google Scholar]
- Engin HB, Gursoy A, Nussinov R, Keskin O. Network-Based Strategies Can Help Mono- and Poly-pharmacology Drug Discovery: A Systems Biology View. Current pharmaceutical design. 2014;20:1201–7. doi: 10.2174/13816128113199990066. [DOI] [PubMed] [Google Scholar]
- Fanger GR, Widmann C, Porter AC, Sather S, Johnson GL, Vaillancourt RR. 14-3-3 proteins interact with specific MEK kinases. The Journal of biological chemistry. 1998;273:3476–83. doi: 10.1074/jbc.273.6.3476. [DOI] [PubMed] [Google Scholar]
- Fantl WJ, Muslin AJ, Kikuchi A, Martin JA, MacNicol AM, Gross RW, Williams LT. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–4. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- Fischer A, Baljuls A, Reinders J, Nekhoroshkova E, Sibilski C, Metz R, Albert S, Rajalingam K, Hekman M, Rapp UR. Regulation of RAF activity by 14-3-3 proteins: RAF kinases associate functionally with both homo- and heterodimeric forms of 14-3-3 proteins. The Journal of biological chemistry. 2009;284:3183–94. doi: 10.1074/jbc.M804795200. [DOI] [PubMed] [Google Scholar]
- Freed E, Symons M, Macdonald SG, McCormick F, Ruggieri R. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science. 1994;265:1713–6. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annual review of pharmacology and toxicology. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Fu H, Xia K, Pallas DC, Cui C, Conroy K, Narsimhan RP, Mamon H, Collier RJ, Roberts TM. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science. 1994;266:126–9. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- Furlanetto RW, Dey BR, Lopaczynski W, Nissley SP. 14-3-3 proteins interact with the insulin-like growth factor receptor but not the insulin receptor. The Biochemical journal. 1997;327(Pt 3):765–71. doi: 10.1042/bj3270765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–40. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–43. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- Harding A, Hancock JF. Ras nanoclusters: combining digital and analog signaling. Cell cycle. 2008;7:127–34. doi: 10.4161/cc.7.2.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold FM. Molecules into cells: specifying spatial architecture. Microbiology and molecular biology reviews : MMBR. 2005;69:544–64. doi: 10.1128/MMBR.69.4.544-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head BP, Patel HH, Insel PA. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochimica et biophysica acta. 2014;1838:532–45. doi: 10.1016/j.bbamem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Ohba Y, Yasuda H. 14-3-3 zeta protein binds to the carboxyl half of mouse wee1 kinase. Biochemical and biophysical research communications. 1997;230:262–5. doi: 10.1006/bbrc.1996.5933. [DOI] [PubMed] [Google Scholar]
- Hou Y, He S, Ma C, Sun M, He H, Gao N. The merged basins of signal transduction pathways in spatiotemporal cell biology. Journal of cellular physiology. 2014;229:287–91. doi: 10.1002/jcp.24449. [DOI] [PubMed] [Google Scholar]
- Irie K, Gotoh Y, Yashar BM, Errede B, Nishida E, Matsumoto K. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–9. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- Janosi L, Li Z, Hancock JF, Gorfe AA. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc Natl Acad Sci U S A. 2012;109:8097–102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Zhao Z. VarWalker: personalized mutation network analysis of putative cancer genes from next-generation sequencing data. PLoS computational biology. 2014;10:e1003460. doi: 10.1371/journal.pcbi.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. The Biochemical journal. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. The EMBO journal. 2000;19:6778–91. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar G, Keskin O, Gursoy A, Nussinov R. Allostery and population shift in drug discovery. Current opinion in pharmacology. 2010;10:715–22. doi: 10.1016/j.coph.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacological reviews. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrath F, Witt J, Sauter T, Kulms D. Identification of new IkappaBalpha complexes by an iterative experimental and mathematical modeling approach. PLoS computational biology. 2014;10:e1003528. doi: 10.1371/journal.pcbi.1003528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelecky B, Saurin AT, Purkiss A, Parker PJ, McDonald NQ. Recognition of an intra-chain tandem 14-3-3 binding site within PKCepsilon. EMBO reports. 2009;10:983–9. doi: 10.1038/embor.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein science : a publication of the Protein Society. 2000;9:10–9. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzu G, Gursoy A, Nussinov R, Keskin O. Exploiting conformational ensembles in modeling protein-protein interactions on the proteome scale. Journal of proteome research. 2013;12:2641–53. doi: 10.1021/pr400006k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine E, Auclair C, Tchertanov L. Allosteric communication across the native and mutated KIT receptor tyrosine kinase. PLoS computational biology. 2012;8:e1002661. doi: 10.1371/journal.pcbi.1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laronga C, Yang HY, Neal C, Lee MH. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. The Journal of biological chemistry. 2000;275:23106–12. doi: 10.1074/jbc.M905616199. [DOI] [PubMed] [Google Scholar]
- Lechtenberg BC, Freund SM, Huntington JA. An ensemble view of thrombin allostery. Biological chemistry. 2012;393:889–98. doi: 10.1515/hsz-2012-0178. [DOI] [PubMed] [Google Scholar]
- Liao J, Omary MB. 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. The Journal of cell biology. 1996;133:345–57. doi: 10.1083/jcb.133.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nussinov R. Flexible cullins in cullin-RING E3 ligases allosterically regulate ubiquitination. The Journal of biological chemistry. 2011;286:40934–42. doi: 10.1074/jbc.M111.277236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Elly C, Yoshida H, Bonnefoy-Berard N, Altman A. Activation-modulated association of 14-3-3 proteins with Cbl in T cells. The Journal of biological chemistry. 1996;271:14591–5. doi: 10.1074/jbc.271.24.14591. [DOI] [PubMed] [Google Scholar]
- Long D, Bruschweiler R. Atomistic kinetic model for population shift and allostery in biomolecules. Journal of the American Chemical Society. 2011;133:18999–9005. doi: 10.1021/ja208813t. [DOI] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein engineering. 1999;12:713–20. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Ma B, Nussinov R. Amplification of signaling via cellular allosteric relay and protein disorder. Proc Natl Acad Sci U S A. 2009;106:6887–8. doi: 10.1073/pnas.0903024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein science : a publication of the Protein Society. 2002;11:184–97. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Tsai CJ, Pan Y, Nussinov R. Why does binding of proteins to DNA or proteins to proteins not necessarily spell function? ACS chemical biology. 2010;5:265–72. doi: 10.1021/cb900293a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkannan S, Awasthi A, Rajasekaran K, Kumar P, Schuldt KM, Bartoszek A, Manoharan N, Goldner NK, Umhoefer CM, Thakar MS. IQGAP1: a regulator of intracellular spacetime relativity. Journal of immunology. 2012;188:2057–63. doi: 10.4049/jimmunol.1102439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, Carter R, Justement LB, Bruckbauer A, Batista FD. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38:461–74. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- McLaughlin RN, Jr., Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–42. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minde DP, Anvarian Z, Rudiger SG, Maurice MM. Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? Molecular cancer. 2011;10:101. doi: 10.1186/1476-4598-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar E, Swamy M, Holzer M, Beck-Garcia K, Worch R, Thiele C, Guigas G, Boye K, Luescher IF, Schwille P, Schubert R, Schamel WW. Cholesterol and sphingomyelin drive ligand-independent T-cell antigen receptor nanoclustering. The Journal of biological chemistry. 2012;287:42664–74. doi: 10.1074/jbc.M112.386045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Puls A, Futter C, Aspenstrom P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. The EMBO journal. 1998;17:149–58. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbs RJ, Graham GJ. Immune regulation by atypical chemokine receptors. Nature reviews. Immunology. 2013;13:815–29. doi: 10.1038/nri3544. [DOI] [PubMed] [Google Scholar]
- Nussinov R. How do dynamic cellular signals travel long distances? Mol Biosyst. 2012;8:22–6. doi: 10.1039/c1mb05205e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. The spatial structure of cell signaling systems. Physical biology. 2013;10:045004. doi: 10.1088/1478-3975/10/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Ma B, Tsai CJ. A broad view of scaffolding suggests that scaffolding proteins can actively control regulation and signaling of multienzyme complexes through allostery. Biochimica et biophysica acta. 2013a;1834:820–9. doi: 10.1016/j.bbapap.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Nussinov R, Ma B, Tsai CJ, Csermely P. Allosteric conformational barcodes direct signaling in the cell. Structure. 2013b;21:1509–21. doi: 10.1016/j.str.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ, Ma B. The underappreciated role of allostery in the cellular network. Annual review of biophysics. 2013c;42:169–89. doi: 10.1146/annurev-biophys-083012-130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ, Xin F, Radivojac P. Allosteric post-translational modification codes. Trends in biochemical sciences. 2012;37:447–55. doi: 10.1016/j.tibs.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Ogihara T, Isobe T, Ichimura T, Taoka M, Funaki M, Sakoda H, Onishi Y, Inukai K, Anai M, Fukushima Y, Kikuchi M, Yazaki Y, Oka Y, Asano T. 14-3-3 protein binds to insulin receptor substrate-1, one of the binding sites of which is in the phosphotyrosine binding domain. The Journal of biological chemistry. 1997;272:25267–74. doi: 10.1074/jbc.272.40.25267. [DOI] [PubMed] [Google Scholar]
- Pallas DC, Fu H, Haehnel LC, Weller W, Collier RJ, Roberts TM. Association of polyomavirus middle tumor antigen with 14-3-3 proteins. Science. 1994;265:535–7. doi: 10.1126/science.8036498. [DOI] [PubMed] [Google Scholar]
- Pandini A, Fornili A, Fraternali F, Kleinjung J. Detection of allosteric signal transmission by information-theoretic analysis of protein dynamics. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:868–81. doi: 10.1096/fj.11-190868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–5. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Pietromonaco SF, Seluja GA, Aitken A, Elias L. Association of 14-3-3 proteins with centrosomes. Blood cells, molecules & diseases. 1996;22:225–37. doi: 10.1006/bcmd.1996.0103. [DOI] [PubMed] [Google Scholar]
- Resendis-Antonio O, Hernandez M, Mora Y, Encarnacion S. Functional modules, structural topology, and optimal activity in metabolic networks. PLoS computational biology. 2012;8:e1002720. doi: 10.1371/journal.pcbi.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuther GW, Fu H, Cripe LD, Collier RJ, Pendergast AM. Association of the protein kinases c-Bcr and Bcr-Abl with proteins of the 14-3-3 family. Science. 1994;266:129–33. doi: 10.1126/science.7939633. [DOI] [PubMed] [Google Scholar]
- Riou P, Kjaer S, Garg R, Purkiss A, George R, Cain RJ, Bineva G, Reymond N, McColl B, Thompson AJ, O’Reilly N, McDonald NQ, Parker PJ, Ridley AJ. 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell. 2013;153:640–53. doi: 10.1016/j.cell.2013.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivalta I, Sultan MM, Lee NS, Manley GA, Loria JP, Batista VS. Allosteric pathways in imidazole glycerol phosphate synthase. Proc Natl Acad Sci U S A. 2012;109:E1428–36. doi: 10.1073/pnas.1120536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Lagree S, Hou Z, Thomson JA, Stewart R, Gasch AP. Integrated module and gene-specific regulatory inference implicates upstream signaling networks. PLoS computational biology. 2013;9:e1003252. doi: 10.1371/journal.pcbi.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Soria P, Camenisch TD. ErbB signaling in cardiac development and disease. Seminars in cell & developmental biology. 2010;21:929–35. doi: 10.1016/j.semcdb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshacharyulu P, Ponnusamy MP, Haridas D, Jain M, Ganti AK, Batra SK. Targeting the EGFR signaling pathway in cancer therapy. Expert opinion on therapeutic targets. 2012;16:15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Han L, Lynch JW. Function of hyperekplexia-causing alpha1R271Q/L glycine receptors is restored by shifting the affected residue out of the allosteric signalling pathway. British journal of pharmacology. 2012;165:2113–23. doi: 10.1111/j.1476-5381.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreif Z, Periwal V. A network characteristic that correlates environmental and genetic robustness. PLoS computational biology. 2014;10:e1003474. doi: 10.1371/journal.pcbi.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn Y, Choi MK, Ahn YY, Lee J, Jeong J. Topological cluster analysis reveals the systemic organization of the Caenorhabditis elegans connectome. PLoS computational biology. 2011;7:e1001139. doi: 10.1371/journal.pcbi.1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldevilla B, Millan CS, Bonilla F, Dominguez G. The TP73 complex network: ready for clinical translation in cancer? Genes, chromosomes & cancer. 2013;52:989–1006. doi: 10.1002/gcc.22095. [DOI] [PubMed] [Google Scholar]
- Song J, Singh M. From hub proteins to hub modules: the relationship between essentiality and centrality in the yeast interactome at different scales of organization. PLoS computational biology. 2013;9:e1002910. doi: 10.1371/journal.pcbi.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Suen TC, McInnes RR, Buchwald M. Association of the TLX-2 homeodomain and 14-3-3eta signaling proteins. The Journal of biological chemistry. 1998;273:25356–63. doi: 10.1074/jbc.273.39.25356. [DOI] [PubMed] [Google Scholar]
- Tinti M, Madeira F, Murugesan G, Hoxhaj G, Toth R, Mackintosh C. ANIA: ANnotation and Integrated Analysis of the 14-3-3 interactome. Database : the journal of biological databases and curation. 2014;2014:bat085. doi: 10.1093/database/bat085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. Journal of molecular biology. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–16. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Kumar S, Ma B, Nussinov R. Folding funnels, binding funnels, and protein function. Protein science : a publication of the Protein Society. 1999a;8:1181–90. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proc Natl Acad Sci U S A. 1999b;96:9970–2. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Ma B, Sham YY, Kumar S, Nussinov R. Structured disorder and conformational selection. Proteins. 2001;44:418–27. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Cox J, Olsen J, Mann M, Frishman D. Phosphorylation variation during the cell cycle scales with structural propensities of proteins. PLoS computational biology. 2013;9:e1002842. doi: 10.1371/journal.pcbi.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Current opinion in structural biology. 2011;21:62–7. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Luo ZJ, Avruch J. Calyculin A-induced vimentin phosphorylation sequesters 14-3-3 and displaces other 14-3-3 partners in vivo. The Journal of biological chemistry. 2000;275:29772–8. doi: 10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Shen YH, Zhu J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene. 2001;20:6331–8. doi: 10.1038/sj.onc.1204777. [DOI] [PubMed] [Google Scholar]
- Vetterkind S, Poythress RH, Lin QQ, Morgan KG. Hierarchical scaffolding of an ERK1/2 activation pathway. Cell communication and signaling : CCS. 2013;11:65. doi: 10.1186/1478-811X-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Molecular and cellular biology. 2000;20:6904–12. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nature genetics. 1998;19:175–8. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- Welz T, Wellbourne-Wood J, Kerkhoff E. Orchestration of cell surface proteins by Rab11. Trends in cell biology. 2014 doi: 10.1016/j.tcb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cellular signalling. 2012;24:826–34. doi: 10.1016/j.cellsig.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–91. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Koo CY, Stebbing J, Giamas G. The dual function of KSR1: a pseudokinase and beyond. Biochemical Society transactions. 2013;41:1078–82. doi: 10.1042/BST20130042. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang H, Masters SC, Wang B, Barbieri JT, Fu H. Residues of 14-3-3 zeta required for activation of exoenzyme S of Pseudomonas aeruginosa. Biochemistry. 1999;38:12159–64. doi: 10.1021/bi991019l. [DOI] [PubMed] [Google Scholar]
- Zhuravlev PI, Papoian GA. Protein functional landscapes, dynamics, allostery: a tortuous path towards a universal theoretical framework. Quarterly reviews of biophysics. 2010;43:295–332. doi: 10.1017/S0033583510000119. [DOI] [PubMed] [Google Scholar]