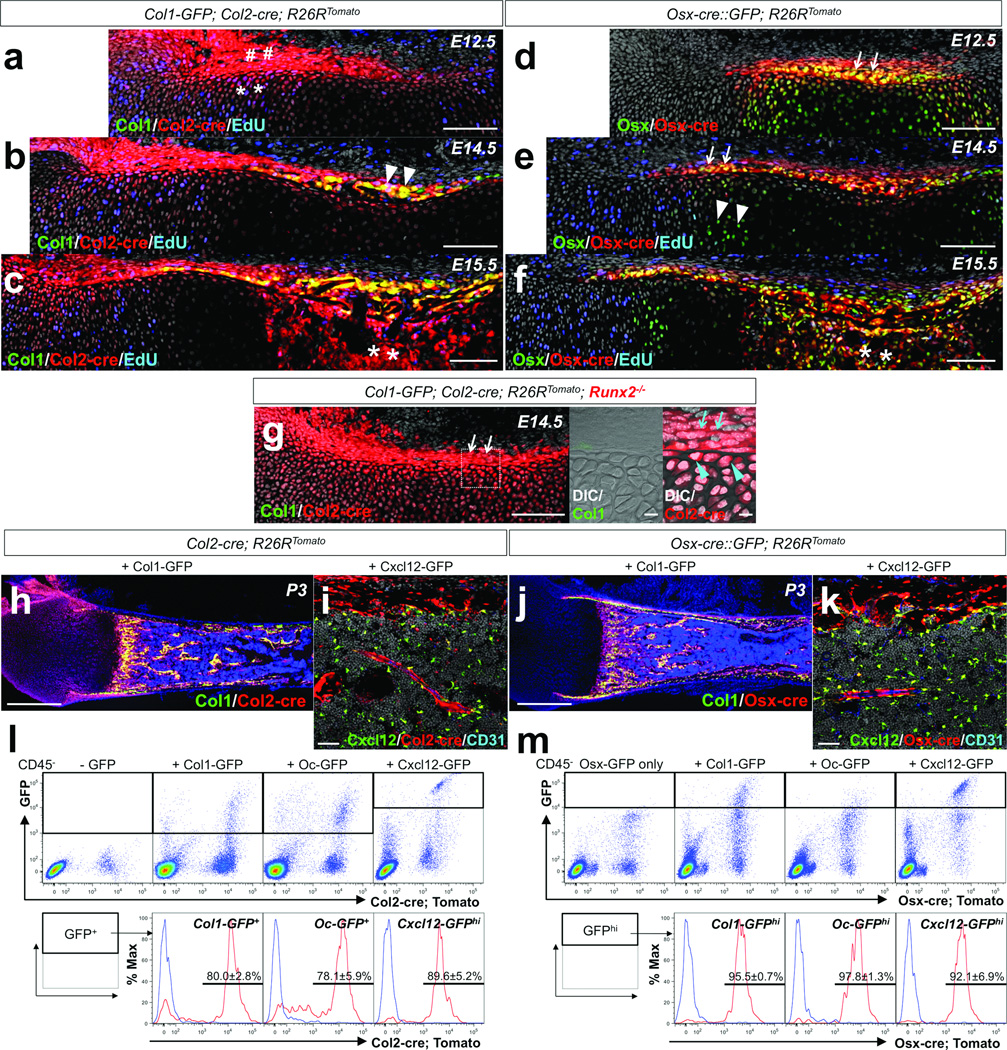
Figure 1. Fate mapping of Col2-cre+ and Osx-cre+ cells during endochondral ossification.
a–c. Fate mapping of Col2-cre+ cells was performed using Col1-GFP; Col2-cre; R26RTomato mice, with EdU administration prior to analysis. (a) Embryonic day 12.5 (E12.5); sharps: Tomato+ perichondrial cells, asterisks: Tomato+ chondrocytes. (b) E14.5; arrowheads: Col1+Tomato+ perichondrial cells. (c) E15.5; asterisks: Tomato+ cells in primary ossification center. Green: EGFP, red: tdTomato, blue: Alexa647, gray: DAPI. Scale bars: 100µm.
d–f. Fate mapping of Osx-cre+ cells was performed using Osx-cre∷GFP; R26RTomato mice, with EdU administration prior to analysis (e,f). (d) E12.5; arrows: Osx+ perichondrium. (e) E14.5; arrows: Osx+Col1− perichondrium, arrowheads: Osx+ prehypertrophic chondrocytes. (f) E15.5; asterisks: Osx+ cells in primary ossification center. Green: EGFP, red: tdTomato, blue: Alexa647, gray: DAPI. Scale bars: 100µm.
g. Fate mapping of Col2-cre+ cells in Runx2-deficient bone anlage at E14.5 was performed using Col1-GFP; Col2-cre; R26RTomato; Runx2−/− mice. Middle and right panels: dotted area of left panel revealing perichondrium. Arrows: Tomato+ perichondrial cells, arrowheads: Tomato+ chondrocytes beneath perichondrium. Green: EGFP, red: tdTomato, gray: DAPI and DIC (differential interference contrast). Scale bars: 100µm (left panel) and 10µm (center and right panels).
h,j. Comparative fate mapping in postnatal day 3 (P3) bones was performed using Col1-GFP; Col2-cre; R26RTomato (h) and Col1-GFP; Osx-cre; R26RTomato (j) mice. Green: EGFP, red: tdTomato, blue: DAPI. Scale bars: 500µm.
i,k. Comparative fate mapping in P3 bones was performed using Cxcl12-GFP; Col2-cre; R26RTomato (i) and Cxcl12-GFP; Osx-cre; R26RTomato (k) mice. Shown are diaphyseal endocortices and bone marrows, stained for CD31. Green: EGFP, red: tdTomato, blue: Alexa633, gray: DAPI. Scale bars: 50µm.
l,m. Flow cytometry analysis was performed using dissociated bone cells harvested at P3. CD45− fraction was gated for GFP. (l) Representative dot plots of cells from Col2-cre; R26RTomato mice that also carry Col1-GFP (left middle panel), Oc-GFP (right middle panel) or Cxcl12-GFP (rightmost panel). Leftmost panel: no GFP control. Lower panels: histograms of the GFP+ fraction developed for Tomato; blue lines: GFP+Tomato control cells. (m) Representative dot plots of cells from Osx-cre∷GFP; R26RTomato mice that also carry Col1-GFP (left middle panel), Oc-GFP (right middle panel) or Cxcl12-GFP (rightmost panel). Leftmost panel: Osx-GFP only control. Lower panels: histograms of the GFPhigh fraction developed for Tomato; blue lines: GFPhighTomato control cells. Upper panels; X-axis: tdTomato, Y-axis: GFP. Lower panels; X-axis: tdTomato. n=3 mice per group. All data are represented as mean ± SD.