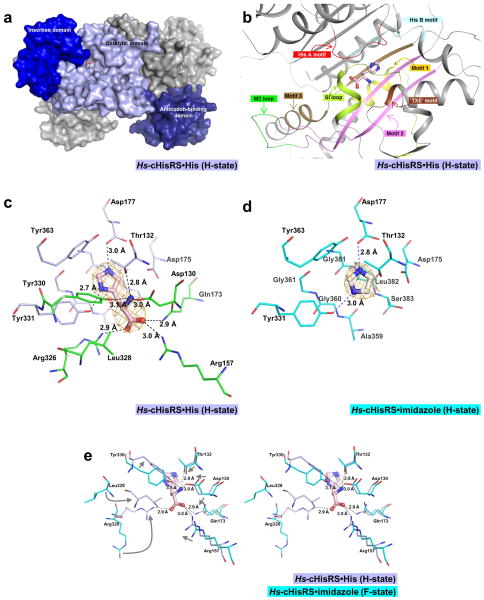
Figure 2. Histidine binding pocket of Hs-cHisRS·His.
a. Surface representation of an Hs-cHisRS·His dimer. The three domains of a monomer are colored in light blue (catalytic domain), blue (insertion domain) and deep blue (anticodon-binding domain). Bound histidine is shown in ball and stick model to indicate the location of the active site. The second monomer is colored gray.
b. Important structural features of HisRS involved in the binding of histidine, with their relative positions with respect to the bound histidine as described in section 3.2. Corresponding sequences of these structural features are annotated in Fig. 1 using the same color scheme.
c. Representative σA-weighted Fobs-Fcalc difference electron density map calculated by omitting histidine is shown at 3σ level. Residues within 4.5Å radius of the bound histidine are shown in sticks. The bound histidine forms eight H-bonds with the protein as indicated by the dash line. Residues that form the imidazole moiety binding site, and are not shifted in positions upon histidine binding, as compared to Fig. 2d, are colored light blue. Residues that are shifted in positions to fully form the histidine binding pocket are colored in green.
d. The σA-weighted Fobs-Fcalc difference electron density map for Hs-cHisRS·imidazole (PDB: 4G84) calculated by omitting imidazole is shown at 3σ level. Residues within 4.5Å radius of the bound imidazole are shown in sticks. Residues that form the imidazole binding site are colored cyan. Compared to Fig. 2c, residues that interact with imidazole moiety do not display significant changes in conformation.
e. Stereo pair showing the superpositions of Hs-cHisRS·imidazole (PDB: 4G84, cyan) and Hs-cHisRS·His (light blue) to indicate the movement of residues (indicated by arrows in the left view) in the active site when the protein changes from F-state to H-state. Significant movements occurred in residues that interact with the amino and carboxylate groups of the bound histidine, mainly through six H-bonds.