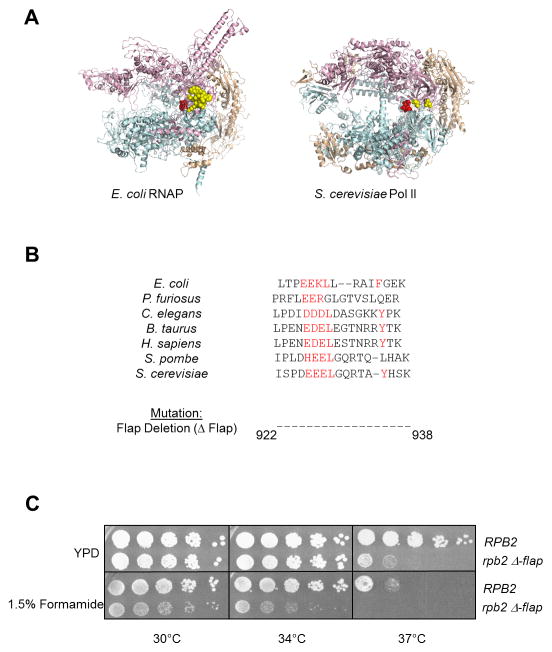
Figure 1. Deletion of the Pol II flap loop in yeast causes phenotypic changes.
(A) Structures of Escherichia coli RNAP (left) and Saccharomyces cerevisiae Pol II (right) showing the β′/Rpb1 subunit in teal, β/Rpb2 subunit in pink, and all other subunits in wheat. The active site residues are shown in red. The E. coli flap loop is highlighted in yellow space-filled rendering, while only the amino-terminal and carboxy-terminal residues of the S. cerevisiae flap loop are shown due to the inherent disordered structure of the loop.
(B) Alignment of the flap loop primary structure across divergent species, with conserved residues highlighted in red. The region corresponding to the S. cerevisiae flap loop (amino acids 922–938 of Rpb2) was deleted to give the rpb2 Δ-flap mutant.
(C) Five-fold serial dilutions of Wt (RPB2) and mutant (rpb2 Δ-flap) Pol II strains are spotted onto YPD +/− 1.5% formamide and incubated at the indicated temperatures for 2–3 days.