Fig. 1.
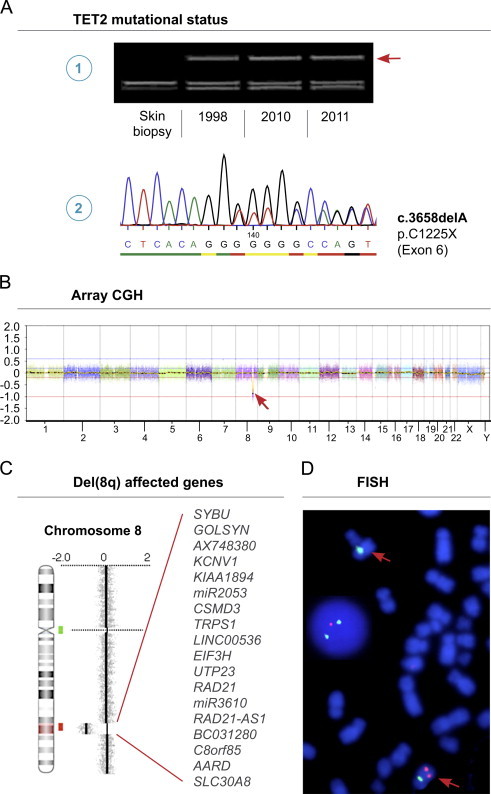
Output data of applied analyses. (A) DGGE analysis showing mutant TET2 being present in all samples as indicated by the heteroduplex band (arrow) (1). Sanger sequencing of the heteroduplex band showing presence of both mutant (del A) and normal alleles (reverse strand shown) (2). (B) High-resolution aCGH analysis at the time of progressive BM failure in 2010 revealing a submicroscopic deletion at chromosome 8 (arrow). The X-axis at the bottom denotes the chromosomal position. (C) Zoom view of genomic profile at chromosome 8 showing the deleted region 8q23.2–24.11 as indicated by red shade on chromosome 8 ideogram. The genes located in the deleted region are listed. Locations of BAC probes for subsequent FISH are indicated (red and green bars). (D) Confirmatory FISH using BAC probe RP11-11A18 at 8q23.3 in the CSMD3 gene (red) and centromeric SE 8 probe (green) showing the deletion on one chromosome 8 (red arrows) in metaphases and interphase nuclei (1R2G signal pattern). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
