Figure 1.
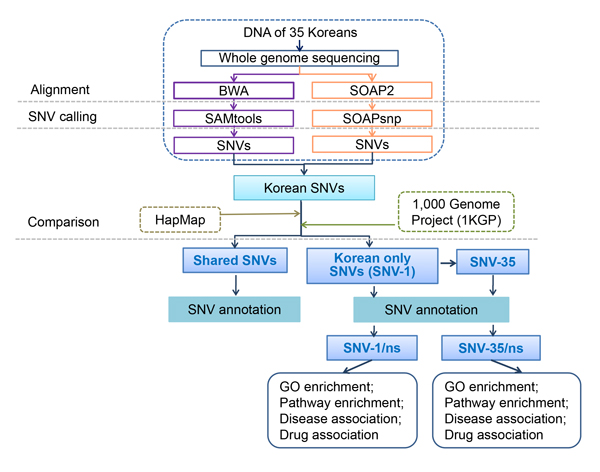
Study design and workflow of this study. The whole genome sequencing data of 35 individuals were used for mapping to the human genome and SNV calling by two pipelines. The overlapped SNVs from the two pipelines were used to represent Korean population and then compared with two references to search for Korean only SNVs and shared SNVs with other populations. Then shared SNVs were annotated. For the Korean only SNVs, two subgroups (SNV-1 and SNV-35) were derived in accordance with the occurrences in the Korean population. The two subgroups of SNVs were then annotated. The non-synonymous SNVs were determined and the corresponding genes were used for enrichment and association analyses.
