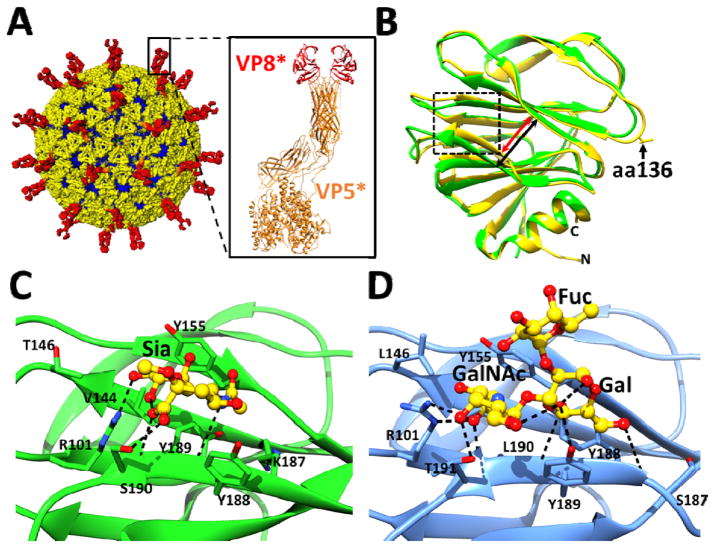
Figure 1.
Rotavirus cryo-EM structure and the crystal structures of cell attachment protein VP8*. (A) The triple layered particle (TLP) is colored with VP4 spikes in red, the VP7 layer in yellow, and the VP6 layer in blue. The cartoon representation of a VP4 spike (PDB ID: 3IYU) is shown with the VP8* domain colored in red and the VP5* domain in orange. (B) Structural overlay of sialidase insensitive P[14] VP8* structure (blue, PDB ID: 4DRV) with VP8* of sialidase-insensitive HR strain Wa (green, PDB ID: 2DWR). The width of the cleft between two twisted β-sheets in the P[14] VP8* is narrower (red arrow) than in the Wa VP8* structure (black arrow). Amino acid 136 is shown in stick and indicated by a black arrow. (C) Interaction between the P[3] VP8* of sialidase sensitive animal strain RRV (PDB ID: 1KQR) and Sia. The P[3] VP8* structure in presented in orange ribbon with the amino acid residues interacting with Sia shown as sticks, and bound Sia is shown as green sticks with oxygen and nitrogen atoms in red and blue, respectively. (D) Interactions between the P[14] VP8* of sialidase insensitive human strain Hal1166 with A-type HBGA. The P[14] VP8* structure in presented in blue ribbon with the amino acid residues interacting with A-HBGA shown as yellow sticks with oxygen and nitrogen atoms colored as in (C). Network of hydrogen bond interactions (dashed lines) are shown.