Abstract
Metabolic engineers develop inexpensive enantioselective syntheses of high-value compounds, but their designs are sometimes confounded by the misfolding of heterologously expressed proteins. Geobacillus stearothermophilus NUB3621 is a readily transformable facultative thermophile. It could be used to express and properly fold proteins derived from its many mesophilic or thermophilic Bacillaceae relatives or to direct the evolution of thermophilic variants of mesophilic proteins. Moreover, its capacity for high-temperature growth should accelerate chemical transformation rates in accordance with the Arrhenius equation and reduce the risks of microbial contamination. Its tendency to sporulate in response to nutrient depletion lowers the costs of storage and transportation. Here, we present a draft genome sequence of G. stearothermophilus NUB3621 and describe inducible and constitutive expression plasmids that function in this organism. These tools will help us and others to exploit the natural advantages of this system for metabolic engineering applications.
Keywords: Geobacillus stearothermophilus, Facultative thermophile, Genome, Expression system
Introduction
Metabolic engineers manipulate microbes in order to convert inexpensive input compounds into valuable outputs. Their designs occasionally fail because foreign proteins, which are necessary for the construction of nonnative metabolic pathways, do not fold properly in host cells. Protein misfolding is all too common because wild-type proteins are marginally stable (average ΔGfolding= −14 kcal/mol (Jaenicke 2000)); apparently modest changes in temperature, amino acid sequence, or chemical environment can cause misfolding or unfolding. Heterologous expression causes foreign proteins and their folding intermediates to interact with high concentrations (300–400 mg/mL) of host protein, sometimes causing aggregation (Baneyx and Mujacic 2004). Many acknowledge these limitations (Woolston et al. 2013), but nevertheless rely heavily upon Escherichia coli and other obligate mesophiles.
Geobacillus stearothermophilus NUB3621 (hereafter called GsNUB3621) offers a general solution to the protein misfolding problem. It belongs to the family Bacillaceae, which diversified to adapt to a variety of very different environments. The functional diversity of this superfamily suggests that natural evolution has already created a versatile set of compatible “parts” that could be artificially mixed and matched without aggregation. GsNUB3621 is a facultative thermophile that achieves balanced growth between 39 and 75 °C (Wu and Welker 1991). It is therefore likely to fold proteins derived from its mesophilic and thermophilic relatives correctly, when propagated at the appropriate temperature. Moreover, others have already demonstrated that GsNUB3621 can serve as a host to direct the evolution of thermostable variants of mesophilic proteins (vide infra).
GsNUB3621 is not the only facultative thermophile, but it is particularly amenable to experimental manipulation. The parent strain, G. stearothermophilus NUB36, was extensively studied by Neil Welker and his colleagues. They developed protocols for the growth, transformation (Wu and Welker 1989), and genetic analysis (Chen et al. 1986) of this strain and created a genetic map (Vallier and Welker 1990). They isolated the GsNUB3621 mutant, which lacked a functioning restriction-modification system. Protoplasts of this strain can be transformed with an efficiency of 107–108 transformants per microgram of DNA (Wu and Welker 1989). Geobacillus thermoglucosidasius (Taylor et al. 2008) and Geobacillus kaustophilus HTA-426 (Suzuki and Yoshida 2012) have also been transformed, but thus far much less efficiently.
Couñago et al. used GsNUB3621 to direct the evolution of thermostable variants of the adenylate kinase gene from Bacillus subtilis (Counago et al. 2006). In this groundbreaking experiment, a mesophilic enzyme was evolved to fold AND function at high temperature. Additionally, Peña et al. have utilized this organism as a vehicle for directed evolution to study the role of protein folding within evolution (Pena et al. 2010). Although it is technically easier to express and fold proteins in E. coli at 37 °C and to assay them ex vivo at higher temperatures (Giver et al. 1998), this approach does not favor mutations that promote proper folding at higher temperatures.
Couñago's feat was even more impressive when one considers the dearth of available genetic tools (plasmids, promoters, and reporter genes) for this nonmodel organism. Such tools must be developed in tandem, as no “positive control” was available to troubleshoot potential constructs. The utility of GsNUB3621 was also limited by the absence of a genome sequence. The genetic map (Vallier and Welker 1990) provides no specific sequences or metabolic pathways. In our experience, degenerate primers based on other Geobacillus genome sequences rarely result in high PCR yields. Here, we present a high-quality draft genome sequence of GsNUB36321 and describe two promoters, one inducible and one constitutive, and two reporter proteins that facilitate the employment of this system for metabolic engineering.
Materials and methods
Materials
All chemicals utilized in this study were from Sigma-Aldrich (St. Louis, MO). DNA oligonucleotides (Table 1) were custom-synthesized by Integrated DNA Technologies (Coraville, IA). All enzymes used for cloning and PCR amplification were purchased from New England Biolabs (Ispwitch, MA). E. coli strain InvaF' (Invitrogen, Grand Island, NY) and custom BioBrick accepting vector pIMBB (Bryksin and Matsumura 2010) were used to clone genes. G. stearothermophilus NUB3621 and shuttle vector pNW33N were provided by the Bacillus Genetic Stock center (Columbus, OH). The Qiaprep Spin Miniprep Kit (Qiagen, Valencia, CA) was used to purify all plasmids (Table 2).
Table 1. Primers used in this study.
| Primer name | Sequence |
|---|---|
| 5′surT for | 5′-cgccgtcggattgcgttccgaagcg-3′ |
| surT EcoRI del for | 5′-agtatgggaaagaatttgcctgcgcgcagaagatggc-3′ |
| surT EcoRI del rev | 5 ′-ccgccatcttctgcgcgcaggcaaattctttcccatactttg-3′ |
| 5′surP rev | 5′-gcgacgcgttcgtaatccatgggcgaacccctctc-3′ |
| 5′agaN for | 5′-gacggaggacaagccatggcaattgtatttgatcc-3′ |
| 3′agaN rev | 5 ′-actagtgcctagccgcatgctagacacc-3′ |
Table 2. Plasmids used in this study.
| Plasmid name | Description | Source |
|---|---|---|
| pNW33N | Geobacillus vector | BGSC, ECE136 |
| pIMBB | BioBrick accepting vector for PCR cloning and assembly | (Bryksin and Matsumura 2010) |
| pIM1638 (GFP-pIMBB) | Accepting vector for surT and agaN fragments | This study |
| pIM241 (agaN-pIMBB) | Source of agaN insert for pIM472 | This study |
| pIM472 (surT-PsurP-agaN-pIMBB) | surT-PsurP-agaN in pIMBB plasmid | This study |
| pIM1708 (surT-PsurP-agaN-pNW33N) | Inducible alpha-galactosidase construct for GsNUB3621 | This study |
| Pldh-sfGFP-pMK-RQ | GeneArt synthesized plasmid containing sfGFP | This study |
| PRHIII-pIDTSmart | IDT minigene construct containing RHIII promoter | This study |
| PRHIII-sfGFP-pIDTSmart | sfGFP cloned downstream of PRHIII promoter | This study |
| pIM1773 (PRHIII-sfGFP-pNW33N) | Constitutive sfGFP construct for GsNUB3621 | This study |
Genome assembly and annotation
Genomic DNA was harvested using Qiagen's DNeasy DNA purification protocol for Gram-positive bacteria. The DNA was then sequenced by whole-genome shotgun sequencing using Illumina technology, generating 37,651,593 100-bp reads. The resulting reads were assembled using ABySS version 1.3.2 and setting the k-mer length to 88 (Simpson et al. 2009). This initial assembly yielded 336 contigs. These contigs were merged to form larger contigs by using iterative BLAST (Altschul et al. 1997) searches to identify regions of overlap between contigs. A database of all of the contigs was generated using the stand-alone version of BLAST. Each of the 50 largest contigs was BLASTed against the database to identify contigs that overlapped the ends. In many instances, multiple contigs were found to overlap with a search input. When possible, these multiple contigs were compared to sequences of other Geobacillus species. Contig placements that matched related Geobacillus species sequences better than the other possible placements were used in the final assembly.
In some instances, contigs that overlapped in the same regions differed by only a few internal nucleotides, most frequently in genomic regions that are present in multiple, nonidentical copies, such as 16S RNA genes. The selection of contigs in our final assembly was thus sometimes arbitrary. The consensus sequence for NUB3621 16S RNA determined here differs from the previously published NUB3621 16S RNA sequence (Goto et al. 2000) by less than 1 % (5 nucleotides out of 1,560), as expected for two 16S RNA copies from the same organism for other Geobacillus species. This consensus sequence was used for all seven 16S RNA copies in the NUB3621 genome although it is unlikely that all are truly identical. This process of identifying contig overlap and extending the largest contigs resulted in ten final contigs containing 3,621,385 bp. The order and orientation of these ten contigs were determined by comparison with the genome of Geobacillus sp. WCH70, which is most similar in sequence to the ten longest of the initial 336 contigs. G. thermoglucosidasius C56-YS93 appeared to be the next most closely related Geobacillus species; its genome sequence supported our current arrangement of contigs. Estimates of gap lengths vary based on whether Geobacillus sp. WCH70 or G thermoglucosidasius C56-YS93 is used as the reference strain, so they are somewhat arbitrary.
The RAST server's online interface was used to annotate the open reading frames (Supplemental Dataset 1); the RAST gene caller was chosen as a basis for annotation. The complete annotation as generated by the RAST server is included. Gaps were set to an arbitrary size of ten nucleotides, which is reflected in the feature start and end positions. For comparison of genes with G. kaustophilus HTA 426, this genome was also annotated using the RAST server and the same parameters. Features that were identical between the two genomes were deleted, duplications within a genome were deleted, and any genes designated as “hypothetical protein” with no additional information were deleted from Supplemental Dataset 2.
Phylogenetic analysis
Maximal unique matches were determined using MUMmer version 3.23 (Kurtz et al. 2004). The minimum match length was set to 19, and both forward and reverse complement matches were set to be reported. Maximal unique matches (MUM) indices were calculated with a published Perl script (Deloger et al. 2009). The tree was generated using SplitsTree4 and choosing the neighbor-joining algorithm (Huson and Bryant 2006). All genome sequences were taken from NCBI database.
Sequence accession numbers
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AOTZ00000000. The version described in this paper is the first version, AOTZ01000000. The deposited version has been annotated by NCBI's Prokaryotic Genome Automatic Annotation Pipeline (PGAAP.)
Bacterial growth conditions
E. coli was grown in either liquid lysogeny broth (LB) media or LB media supplemented with 1.5 % (w/v) agar at a temperature of 37 °C. GsNUB3621 was grown in modified LB (mLB) media (Chen et al. 1986), which also contains 1.05 mM nitrilotriacetic acid, 0.59 mM MgSO4·7H2O, 0.91 mM CaCl2·2H2O, 0.04 mM FeSO4·7H2O, and 1.5 % (w/v) agar as needed. GsNUB3621 was grown at 60 °C unless otherwise noted. When grown in liquid culture, GsNUB3621 was grown with minimal volumes to maximize aeration (20 mL in a 250-mL flask.) The LB media was supplemented with 100 μg/mL ampicillin for selection of pIMBB-based plasmids in E. coli or 34 μg/mL of chloramphenicol for selection of pNW33N-based plasmids. The mLB medium was augmented with 7 μg/mL of chloramphenicol for selection of GsNUB3621 transformed with pNW33N plasmids.
GsNUB3621 transformation
GsNUB3621 protoplasts were transformed as described previously (Wu and Welker 1989) but with several modifications. Cultures were grown to saturation overnight, diluted 1/40, and propagated for 4.5 h with shaking at 60 °C until the cell density reached an OD 600 of approximately 1.8. For each transformation, 2 mL of log-phase culture was harvested in a microcentrifuge for 1 min at 5,000 rpm. The cells were resuspended in 500 μL of protoplasting media (mLB with 10 %w/v lactose and 10 mM MgCl2·7H2O); 10 μL of 1 mg/mL lysozyme were added to each and incubated for 20 min at 37 °C in a water bath. The conversion of whole cells to protoplasts was verified by microscopy.
The protoplasts were then diluted with 500 μL of protoplasting media, harvested by centrifugation (5 min at 800g), and resuspended in 100 μL of protoplasting media. One microgram of the appropriate plasmid DNA (100 ng/μL × 10 μL) was added to the washed protoplasts; 900 μL of freshly prepared PEG solution (40 %w/v PEG 6000 in protoplasting media) was subsequently added. The protoplast/DNA/PEG mixtures were then incubated for 2 min at 50 °C with shaking (130 rpm), harvested by centri-fugation for 5 min at 800g, resuspended in 100 μL of protoplasting media, and incubated for an hour at 50 °C with shaking at 130 rpm. The mixtures were then spread on regeneration plates (protoplasting media with 20 mM CaCl2·2H2O, 0.8 % agar, and 7 μg/mL chloramphenicol), incubated for 12 h at 50 °C, then at 60 °C until the appearance of colonies. At the first sign of growth, the colonies were replicaplated onto fresh mLB plates supplemented with 7 μg/mL of chloramphenicol and incubated at 60 °C overnight.
Cloning of surT-PsurP-agaN-pNW33N and PRHIII-sfGFP-pNW33N
The agaN reporter gene was PCR-amplified from NUB3621 genomic DNA and blunt-end-cloned into an EcoRV-cut pIMBB (Table 3) accepting vector (Bryksin and Matsumura 2010). The regulatory region upstream of the surT promoter to the surP start codon was PCR-amplified from GsNUB3621 genomic DNA. An EcoRI site within this region was eliminated by introducing a synonymous single base pair change in two separate PCRs that were then joined in an overlap extension PCR. The surT-PsurP regulatory region was then joined to agaN in the pIMBB cloning plasmid by cutting the surT-PsurP PCR product with NspI and NcoI, cutting GFP-pIMBB with SphI and SpeI, and cutting agaN-pIMBB with NcoI and SpeI and ligating the three fragments together. The surT-PsurP-agaN fragment was then cloned from the pIMBB vector into pNW33N by cutting both plasmids with EcoRI and PstI. The plasmid was deposited into the Addgene repository (ID 44009).
Table 3. Contigs in GsNUB3621 draft genome.
| Contig number | Length (bp) | Starta | Stop | Estimated gapb (bp) |
|---|---|---|---|---|
| 1 | 159,532 | 105368 | 267177 | 17,644 |
| 2 | 615,890 | 284821 | 996044 | 3,849 |
| 3 | 411,492 | 999893 | 1424910 | 34,581 |
| 4 | 537,992 | 1458591 | 1812705 | 8,684 |
| 5 | 361,973 | 1821389 | 2055928 | 16,933 |
| 6 | 831,535 | 2072861 | 2838732 | 5,278 |
| 7 | 14,434 | 2844010 | 2847735 | 25,660 |
| 8 | 103,904 | 2873395 | 2969136 | 1,853 |
| 9 | 441,948 | 2970989 | 3415682 | 166 |
| 10 | 142,685 | 3415848 | 91412 | 13,956 |
The start and stop values represent the approximate mapping locations in Geobacillus sp. WCH70. A start value of 105368 means that the most 5′ BLAST hit matched the region starting at base pair 105368 in Geobacillus sp. WCH70
Gap estimate is for the gap following each contig
The sfGFP gene was synthesized by GeneArt to reflect the codon bias of G. stearothermophilus (Nakamura et al. 2000); internal EcoRI (nucleotide 669) and NdeI (nucleotide 231) sites were changed to synonymous codons. The BioBrick prefix was added to the 5′ end and a BioBrick suffix was added to the 3′ end. An NcoI site was added at the start codon and a glycine sequence was added in the second amino acid position to correct a frameshift that would otherwise have been caused by the addition of the NcoI site. The ribonuclease H III promoter was synthesized by IDT as a minigene construct. Restriction endonucleases NcoI and PstI were used to subclone the sfGFP gene into pIDTSmart downstream of PRHIII. EcoRI and PstI were then used to subclone the PRHIII-sfGFP cassette into pNW33N to create PRHIII-sfGFP-pNW33N (Addgene 52217).
Alpha-galactosidase assays
G. stearothermophilus NUB3621 assays were carried out as described previously (Beutler and Kuhl 1972; Talbot and Sygusch 1990) with slight modifications. Reactions containing 850 μL of supernatant, 50 μL of 1 M Tris pH 7.6, and 100 μL of 40 mM 4-methylumbelliferyl-alpha-D-galactopyranoside (4-MU) were incubated at 55 °C; 10 μL aliquots were removed at 5 minute intervals and added to 100 μL of 0.2 N NaOH in 96 well, white microtiter plates to quench the reaction. Fluorescence was measured in a SpectraMax M5 plate reader using an excitation wavelength of 355 nm and an emission wavelength of 460 nm. The photomultiplier automatic setting was used and the slit widths were set to the defaults (9 nm for excitation and 15 nm for emission.)
sfGFP assays
GsNUB3621 carrying PRHIII-sfGFP-pNW33N was propagated overnight to saturation. The cells were harvested by cen-trifugation at 3,500 rpm for 15 min; the cells were resuspended in 2 mL of 50 mM Tris pH 8.0, 10 mM EDTA, 2 mg/mL lysozyme and incubated at 37 °C for 30 min. The cell debris was removed by centrifugation for 2.5 min at 7,000 rpm; 100 μL aliquots of the supernatant were transferred to 96-well microtiter plates and assayed. Fluorescence was measured in a SpectraMax M5 plate reader using an excitation wavelength of 470 nm and a cutoff value of 495 nm. The photomultiplier automatic setting was used, and the slit widths were set to the defaults (9 nm for excitation and 15 nm for emission.)
Results
Genome sequence of GsNUB3621
Genome sequences have become de rigueur for metabolic engineering. Couñago et al., for example, worked without a genome sequence, which meant that they had to sequence their knockout target gene in the GsNUB3621 chromosome and show that it was essential (Couñago and Shamoo 2005). We used the Illumina method to sequence the GsNUB3621 genome. Assembly of the reads resulted in ten ordered contigs (Table 1), an “improved high-quality draft genome” (Chain et al. 2009). Scaffolding was completed by aligning the contigs against a related genome sequence, that of Geobacillus sp. WCH70. We emphasize that the gap estimates could differ substantially from the true gap sizes. In some instances, the first or last several kilobases of a contig had no sequence identity to Geobacillus sp. WCH70. Thus, the estimated “start” or “end” positions for some contigs correlate to positions a few kilobases within the contig, rather than the first or last nucleotide of the contig. We tried to resolve these gaps via targeted sequencing but were unable to PCR amplify them. The gaps might be longer than we estimate or contain repetitive sequences.
To verify the current order and orientation of the contigs, we also compared the contigs to the genome of G. thermoglucosidasius C56-YS93 (not shown). The BLAST hits against the G. thermoglucosidasius C56-YS93 genome confirm the general order and orientation of the contigs, although the gap estimates vary, and synteny within contigs is not always conserved between Geobacillus sp.WCH70 and G. thermoglucosidasius C56-YS93. We also attempted to validate this scaffold by comparison with the existing GsNUB36 genetic map (Vallier and Welker 1990). Although we cannot be certain of the genetic loci that correspond to the phenotypes used to generate the map, we identified plausible candidate loci for many of the phenotypes, and our assignments were generally in concordance with the map (not shown).
The genome sequence was annotated using the RAST server (Aziz et al. 2008). The RAST gene caller identified 3,929 features (annotations, listed in Supplemental Dataset 1) with an estimated 8 features possibly missing from the sequence. The algorithm classified 47 % of these features into RAST gene subsystems (Fig. 1); it was unable to predict the functions of the majority of genes. The percentage of subsystem coverage, as well as the subsystem feature counts, is similar to that of G. kaustophilus HTA426 and Geobacillus thermodenitrificans NG80-2, which were already present in the RAST database's SEED viewer (Overbeek et al. 2005). The features identified were then compared to those of the more extensively studied G. kaustophilus HTA426 genome (Takami et al. 2004a; Takami et al. 2004b). This comparison revealed 842 different features (Supplemental Dataset 2) between the two organisms (440 uniqueto GsNUB3621 and 402 unique to HTA426). Among the unique genes, G. kaustophilus HTA426 includes multiple restriction endonucleases, while GsNUB3621 has none. We hypothesize that this difference explains why the latter is more amenable to transformation.
Fig. 1.
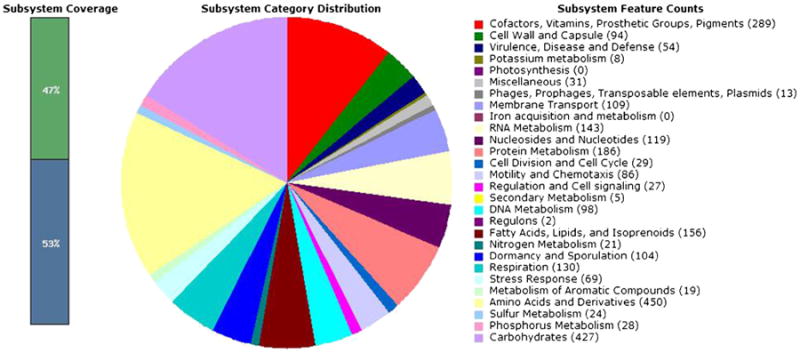
The RAST server (Aziz et al. 2008) was used to annotate the open reading frames of GsNUB3621. Of the 3,832 features that were identified, 47 % fell into known gene categories. The distribution of their functions is shown
The phylogenetic position of GsNUB3621 was somewhat uncertain (Studholme et al. 1999; Zeigler 2005), so we compared our genome sequence to that of other Geobacillus genomes (Coorevits et al. 2011). The MUM index (MUMi) is a computationally tractable method of estimating species relatedness utilizing whole-genome data (Deloger et al. 2009). Briefly, MUMi utilizes the maximal unique matches (MUM) between two species as a measure of relatedness. MUMi values between GsNUB3621 and the 11 available Geobacillus whole-genome sequences (Geobacillus sp. WCH70, accession number NC_012793.1; G. thermoglucosidasius C56-YS93, accession number NC_015660.1; G. kaustophilus HTA426, accession number NC_006510.1 (Takami et al. 2004b); G. thermodenitrificans NG80-2, accession number NC_009328.1 (Feng et al. 2007); Geobacillus thermoleovorans CCB_US3_UF5, accession number NC_016593.1; Geobacillus sp. Y4.1MC1, accession number NC_014650.1; Geobacillus sp. Y412MC52, accession number NC_014915.1; Geobacillus sp. Y412MC61, accession number NC_013411.1; Geobacillus sp. C56-T3, accession number NC_014206.1; Geobacillus sp. GHH01, accession number NC_020210.1; and Geobacillus thermoglucosidans TNO-09.020, accession number NZ_CM001483.1 (Zhao et al. 2012)) were calculated and used to construct a phylogenetic tree (Fig. 2). Our GsNUB3621 sequence is not identical to any of the other sequenced genomes. It remains unclear whether GsNUB3621 is closely related to other G. stearothermophilus strains, including the type strain, as none have yet been fully sequenced.
Fig. 2.
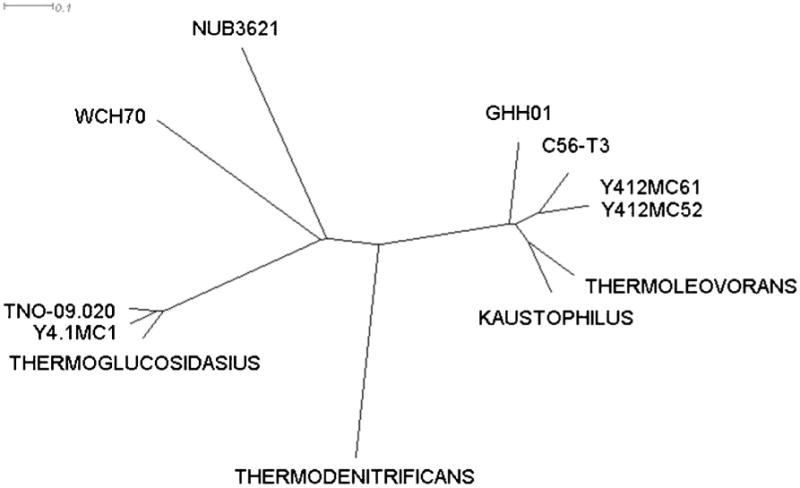
The maximal unique matches index (Coorevits et al. 2011) was used to compare the GsNUB3621 genome sequence with those of other Geobacillus species (namely, Geobacillus sp. WCH70, Geobacillus thermoglucosidasius C56-YS93, Geobacillus kaustophilus HTA426, Geobacillus thermodenitrificans NG80-2, Geobacillus thermoleovorans CCB_US3_UF5, Geobacillus sp. Y4.1MC1, Geobacillus sp. Y412MC52, Geobacillus sp. Y412MC61, Geobacillus sp. C56-T3, Geobacillus sp. GHH01, and Geobacillus thermoglucosidans TNO-09.020). The phylogenetic separation between Geobacillus sp. Y412MC52 and Geobacillus sp. Y412MC61 can only be detected at higher resolutions
Inducible and constitutive expression of reporter genes in GsNUB3621
Couñago et al. integrated a foreign gene into an existing operon within the GsNUB3621 chromosome (Couñago and Shamoo 2005). A plasmid would enable more efficient transformations, which are essential for in vitro mutagenesis and recombination protocols. An inducible expression system would facilitate the heterologous expression of toxic genes. We constructed our inducible expression vector, surT-PsurP-agaN-pNW33N, by combining parts that were developed or characterized by others. The parent vector for this plasmid, pNW33N, contains a chloramphenicol acetyltransferase gene that confers chloramphenicol resistance upon both E. coli and GsNUB3621 at elevated temperatures (De Rossi et al. 1994). This plasmid is fortuitously compatible with the BioBrick standard, a system of standardized restriction sites that facilitate the combinatorial assembly of multicomponent biological devices (Shetty et al. 2008).
The sucrose utilization operon regulatory region (Li and Ferenci 1997) consists of a sucrose phosphotransferase gene surP that is regulated by the surT gene product. The surT antiterminator is believed to bind to palindromic surR region, allowing sucrose-induced transcription from the surP promoter (Fig. 3). Our genome sequence data indicated that GsNUB3621 possesses two genes that encode alpha-galactosidases. One matched the previously published GsNUB3621 agaN sequence (Fridjonsson et al. 1999), while the other was more distantly related. The former homologue was PCR-amplified from GsNUB3621 and cloned downstream of surT-PsurP, thereby completing our surT-PsurP-agaN-pNW33N expression vector. GsNUB3621 was transformed via protoplast transformation (Materials and methods) and assayed for activity both in the presence and absence of sucrose. The alpha-galactosidase activity in the supernatant increased fivefold when cultures were grown in sucrose (Fig. 4a). Control cells carrying only the empty pNW33N plasmid exhibited little alpha-galactosidase activity, which suggests that the chromosomal copy is not expressed under these conditions and that our inducible promoter is somewhat leaky.
Fig. 3.
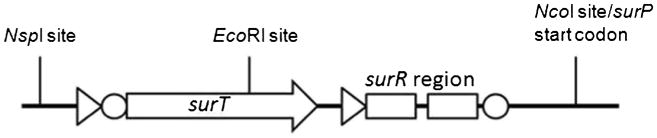
The regulatory region of the sucrose utilization operon is schematized. Triangles denote promoters while circles represent Shine-Dalgarno sequences. The surT gene encodes an antiterminator that is thought to bind the surR region, allowing transcription from the surP promoter. The region was amplified via PCR; the EcoRI site within surT was incompatible with the BioBrick cloning standard, so it was eliminated by site-directed mutagenesis of a single base to effect a synonymous substitution. An NcoI site was created at the surP start codon; the region was cloned into the E. coli/GsNUB3621 shuttle plasmid pNW33N with restriction enzymes NcoI, NspI, and SphI
Fig. 4.
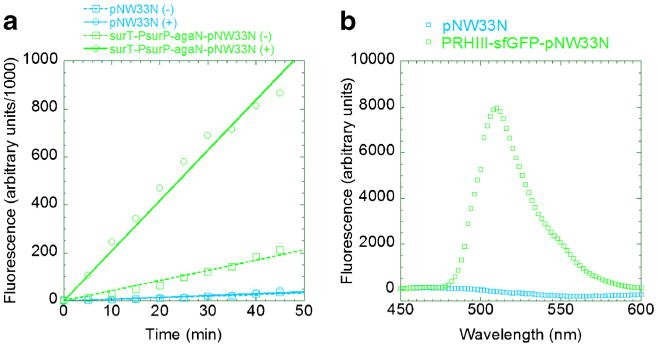
The inducible surP promoter (a) and constitutive ribonuclease HIII promoter (b) were used to express reporter proteins alpha-galactosidase (AgaN) and superfolding green fluorescent protein (sfGFP), respectively. a GsNUB3621 were transformed with the empty E. coli/GsNUB3621 shuttle vector pNW33N (blue) or surT-PsurP-agaN-pNW33N (green). The transformants were propagated overnight in modified LB medium, in the presence (solid lines) or absence (dotted lines) of sucrose. The supernatants were reacted with 4-methylumbelliferyl-alpha-D-galactopyranoside. Aliquots were quenched in sodium hydroxide; the alpha-galactosidase activity (increase in fluorescence over time) was measured in a spectrofluorimeter. b GsNUB3621 was transformed with the empty pNW33N vector (blue) or expression vector PRHIII-sfGFP--pNW33N (green). The transformants were propagated overnight in mLB medium. The cells were harvested by centrifugation, resuspended in buffer, and lysed by lysozyme-catalyzed hydrolysis of their cell walls. The fluorescence spectra were measured; the values were adjusted by subtracting the fluorescence of a blank (fresh mLB)
We looked through our sequence data to identify constitutive promoters. Among those considered, we focused on the promoter for ribonuclease H III (PRHIII) gene because its −10 and −35 regions most closely matched the consensus. We coupled this promoter to the superfolding green fluorescent protein (sfGFP), a thermostable variant of GFP (Pedelacq et al. 2006). Fluorescence was detected in cell extracts derived from GsNUB3621 transformed with PRHIII-sfGFP-pNW33N, but not in control extracts with the empty vector (pNW33N) control (Fig. 4b).
In addition to the surT-PsurP-agaN-pNW33N and PRHIII-sfGFP-pNW33N plasmids, we cloned and tested several others that did not work as well. We coupled either agaN or sfGFP to the following promoters: T5 (Bujard et al. 1987), T7 (Studier and Moffatt 1986), tac (de Boer et al. 1983), spac (Yansura and Henner 1984), and the promoter from the chlor-amphenicol resistance gene present in pNW33N. We were unable to construct PSpac-agaN-pNW33N and PRHIII-agaN-pNW33N, which suggested to us that high levels of AgaN expression are toxic to E. coli. We constructed PSpac-sfGFP-pNW33N, lacI-PT5-agaN-pNW33N, and lacI-Ptac-agaN-pNW33N in E. coli but were unable to obtain viable GsNUB3621 transformants. We transformed GsNUB3621 with Pcat-agaN-pNW33N and Pcat-sfGFP-pNW33N, but we did not observe any reporter protein expression. The lacI-PT7-agaN-pNW33N plasmid apparently caused GsNUB3621 to express modest amounts of AgaN, less than twofold change over control cells carrying the empty pNW33N plasmid (data not shown).
Discussion
GsNUB3621 as source of robust proteins
Proteins that are robust to mutation tend to be more evolvable. Mutational robustness and conformational stability are correlated (Bloom et al. 2006); thermophilic bacteria are full of thermostable enzymes, but these might not be good starting points for directed evolution because few are active at mesophilic temperatures. In contrast, GsNUB3621 can grow at temperatures between 39 and 75 °C so its proteins would probably be good starting points for the directed evolution of variants with novel catalytic functions (promiscuous or substrate ambiguous activities) at physiological temperatures. The GsNUB3621 genome sequence could be used to identify candidate genes for PCR amplification, cloning in an E. coli expression vector, and subsequent directed evolution in that mesophilic host. Alternatively, it should be relatively easy to assay GsNUB3621 cell extracts for desired catalytic activities, to purify those thermostable activities via classical biochemical fractionation, and to identify the gene through the mass or amino acid sequence of its product.
GsNUB3621 as a host strain for metabolic engineering applications
We developed an expression plasmid for GsNUB3621 and sequenced its genome, so that we and others can use this organism as a vehicle for the directed evolution of robust variants of mesophilic proteins. GsNUB3621 should also be a good host for metabolic engineering applications. Others have already shown that a related species, G. themoglucosidasius, is useful for biofuel production (Taylor et al. 2008). Enzyme-catalyzed reactions follow the Arrhenius equation, which predicts that reaction rates should increase exponentially with temperature (50–100 % faster for each 10 °C). Most enzymes denature above their optimum temperatures, but robust pathways within robust organisms are likely to be much faster and more efficient. Furthermore, bioreactors are much less likely to become contaminated at high temperatures. GsNUB3621 sporulates under starvation conditions (Wu and Welker 1991). G. stearothermophilus endospores are commonly used to test autoclaves and other disinfection protocols, so we tentatively expect that GsNUB3621 endospores will similarly be resilient. The capacity of endospores to persist indefinitely without refrigeration will save energy and simplify the transport of engineered strains to remote locations. The tools described here allow us and others to exploit the natural advantages GsNUB3621 for metabolic engineering applications.
Supplementary Material
Acknowledgments
We thank the Bacillus Genetic Stock Center for GsNUB3621, the pNW33N plasmid, and their protocols. We also thank the Emory Integrated Genomics Core for their service and Paul Doetsch for the use of his spectrofluorimeter. This work was supported by the National Institute of General Medicine at the National Institutes for Health. KB and IM were supported by R01 GM086824 and KB was also supported by 5T32GM008490-19.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00253-014-5746-z) contains supplementary material, which is available to authorized users.
Contributor Information
Kristen Blanchard, Department of Biochemistry, Emory University School of Medicine, 1510 Clifton Road NE, room 4119, Atlanta, GA 30322, USA.
Srebrenka Robic, Department of Biology, Agnes Scott College, 141 E. College Ave., Decatur, GA 30030, USA.
Ichiro Matsumura, Email: imatsum@emory.edu, Department of Biochemistry, Emory University School of Medicine, 1510 Clifton Road NE, room 4119, Atlanta, GA 30322, USA.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22(11):1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- Beutler E, Kuhl W. Purification and properties of human alpha-galactosidases. J Biol Chem. 1972;247(22):7195–7200. [PubMed] [Google Scholar]
- Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci U S A. 2006;103(15):5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48(6):463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle MT, Dobberstein B. A T5 promoter-based transcription-translation system for the analysis of proteins in vitro and in vivo. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, Cole JR, Ding Y, Dugan S, Field D, Garrity GM, Gibbs R, Graves T, Han CS, Harrison SH, Highlander S, Hugenholtz P, Khouri HM, Kodira CD, Kolker E, Kyrpides NC, Lang D, Lapidus A, Malfatti SA, Markowitz V, Metha T, Nelson KE, Parkhill J, Pitluck S, Qin X, Read TD, Schmutz J, Sozhamannan S, Sterk P, Strausberg RL, Sutton G, Thomson NR, Tiedje JM, Weinstock G, Wollam A, Detter JC. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326(5950):236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Wojcik SF, Welker NE. Genetic analysis of Bacillus stearothermophilus by protoplast fusion. J Bacteriol. 1986;165(3):994–1001. doi: 10.1128/jb.165.3.994-1001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coorevits A, Dinsdale AE, Halket G, Lebbe L, De Vos P, Van Landschoot A, Logan NA. Taxonomic revision of the genus Geobacillus: emendation of Geobacillus, G. stearothermophilus, G. jurassicus, G. toebii, G. thermodenitrificans and G. thermoglucosidans (nom. corrig., formerly ‘thermoglucosidasius’); transfer of Bacillus thermantarcticus to the genus as G. thermantarcticus comb. nov.; proposal of Caldibacillus debilis gen. nov., comb. nov.; transfer of G. tepidamans to Anoxybacillus as A. tepidamans comb. nov.; and proposal of Anoxybacillus caldiproteolyticus sp. nov. Int J Syst Evol Microbiol. 2011;62(Pt 7):1470–1485. doi: 10.1099/ijs.0.030346-0. [DOI] [PubMed] [Google Scholar]
- Couñago R, Shamoo Y. Gene replacement of adenylate kinase in the gram-positive thermophile Geobacillus stearothermophilus disrupts adenine nucleotide homeostasis and reduces cell viability. Extremophiles. 2005;9(2):135–144. doi: 10.1007/s00792-004-0428-x. [DOI] [PubMed] [Google Scholar]
- Couñago R, Chen S, Shamoo Y. In vivo molecular evolution reveals biophysical origins of organismal fitness. Mol Cell. 2006;22(4):441–449. doi: 10.1016/j.molcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- de Boer HA, Comstock LJ, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi E, Brigidi P, Welker NE, Riccardi G, Matteuzzi D. New shuttle vector for cloning in Bacillus stearothermophilus. Res Microbiol. 1994;145(8):579–583. doi: 10.1016/0923-2508(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Deloger M, El Karoui M, Petit MA. A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol. 2009;191(1):91–99. doi: 10.1128/JB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, Tang Y, Liu X, Han W, Peng X, Liu R, Wang L. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci U S A. 2007;104(13):5602–5607. doi: 10.1073/pnas.0609650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridjonsson O, Watzlawick H, Gehweiler A, Mattes R. Thermostable alpha-galactosidase from Bacillus stearothermophilus NUB3621: cloning, sequencing and characterization. FEMS Microbiol Lett. 1999;176(1):147–153. doi: 10.1111/j.1574-6968.1999.tb13655.x. [DOI] [PubMed] [Google Scholar]
- Giver L, Gershenson A, Freskgard PO, Arnold FH. Directed evolution of a thermostable esterase. Proc Natl Acad Sci U S A. 1998;95(22):12809–12813. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Omura T, Hara Y, Sadaie Y. Application of the partial 16S rDNA sequence as an index for rapid identification of species in the genus Bacillus. J Gen Appl Microbiol. 2000;46(1):1–8. doi: 10.2323/jgam.46.1. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jaenicke R. Stability and stabilization of globular proteins in solution. J Biotechnol. 2000;79(3):193–203. doi: 10.1016/s0168-1656(00)00236-4. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ferenci T. Gene organisation and regulatory sequences in the sucrose utilisation cluster of Bacillus stearothermophilus NUB36. Gene. 1997;195(2):195–200. doi: 10.1016/s0378-1119(97)00139-x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28(1):292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24(1):79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Pena MI, Davlieva M, Bennett MR, Olson JS, Shamoo Y. Evolutionary fates within a microbial population highlight an essential role for protein folding during natural selection. Mol Syst Biol. 2010;6:387. doi: 10.1038/msb.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J Biol Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme DJ, Jackson RA, Leak DJ. Phylogenetic analysis of transformable strains of thermophilic Bacillus species. FEMS Microbiol Lett. 1999;172(1):85–90. doi: 10.1111/j.1574-6968.1999.tb13454.x. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Yoshida K. Genetic transformation of Geobacillus kaustophilus HTA426 by conjugative transfer of host-mimicking plasmids. J Microbiol Biotechnol. 2012;22(9):1279–1287. doi: 10.4014/jmb.1203.03023. [DOI] [PubMed] [Google Scholar]
- Takami H, Nishi S, Lu J, Shimamura S, Takaki Y. Genomic characterization of thermophilic Geobacillus species isolated from the deepest sea mud of the Mariana Trench. Extremophiles. 2004a;8(5):351–356. doi: 10.1007/s00792-004-0394-3. [DOI] [PubMed] [Google Scholar]
- Takami H, Takaki Y, Chee GJ, Nishi S, Shimamura S, Suzuki H, Matsui S, Uchiyama I. Thermoadaptation trait revealed by the genome sequence of thermophilic Geobacillus kaustophilus. Nucleic Acids Res. 2004b;32(21):6292–6303. doi: 10.1093/nar/gkh970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot G, Sygusch J. Purification and characterization of thermostable beta-mannanase and alpha-galactosidase from Bacillus stearothermophilus. Appl Environ Microbiol. 1990;56(11):3505–3510. doi: 10.1128/aem.56.11.3505-3510.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Esteban CD, Leak DJ. Development of a versatile shuttle vector for gene expression in Geobacillus spp. Plasmid. 2008;60(1):45–52. doi: 10.1016/j.plasmid.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Vallier H, Welker NE. Genetic map of the Bacillus stearothermophilus NUB36 chromosome. J Bacteriol. 1990;172(2):793–801. doi: 10.1128/jb.172.2.793-801.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolston BM, Edgar S, Stephanopoulos G. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng. 2013 doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Welker NE. Protoplast transformation of Bacillus stearothermophilus NUB36 by plasmid DNA. J Gen Microbiol. 1989;135(5):1315–1324. doi: 10.1099/00221287-135-5-1315. [DOI] [PubMed] [Google Scholar]
- Wu L, Welker NE. Temperature-induced protein synthesis in Bacillus stearothermophilus NUB36. J Bacteriol. 1991;173(15):4889–4892. doi: 10.1128/jb.173.15.4889-4892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yansura DG, Henner DJ. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984;81(2):439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR. Application of a recN sequence similarity analysis to the identification of species within the bacterial genus Geobacillus. Int J Syst Evol Microbiol. 2005;55(Pt 3):1171–1179. doi: 10.1099/ijs.0.63452-0. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Caspers MP, Abee T, Siezen RJ, Kort R. Complete genome sequence of Geobacillus thermoglucosidans TNO-09.020, a thermophilic sporeformer associated with a dairy-processing environment. J Bacteriol. 2012;194(15):4118. doi: 10.1128/JB.00318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


