Fig. 3.
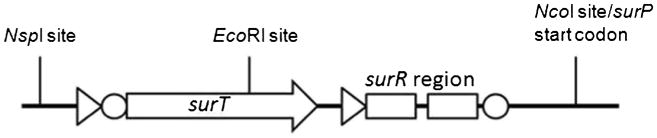
The regulatory region of the sucrose utilization operon is schematized. Triangles denote promoters while circles represent Shine-Dalgarno sequences. The surT gene encodes an antiterminator that is thought to bind the surR region, allowing transcription from the surP promoter. The region was amplified via PCR; the EcoRI site within surT was incompatible with the BioBrick cloning standard, so it was eliminated by site-directed mutagenesis of a single base to effect a synonymous substitution. An NcoI site was created at the surP start codon; the region was cloned into the E. coli/GsNUB3621 shuttle plasmid pNW33N with restriction enzymes NcoI, NspI, and SphI
