Abstract
Background
The procoagulant state in cancer increases the thrombotic risk, but also supports tumor progression. To investigate the molecular mechanisms controlling cancer and hemostasis, we conducted a case-control study of genotypic and phenotypic variables of the tissue factor (TF) pathway of coagulation in breast cancer.
Methods
366 breast cancer patients and 307 controls were genotyped for SNPs (n = 41) in the F2, F3 (TF), F5, F7, F10, TFPI and EPCR genes, and assayed for plasma coagulation markers (thrombin generation, activated protein C (APC) resistance, D-dimer, antithrombin, protein C, protein S, and TF pathway inhibitor (TFPI)). Associations with breast cancer were evaluated using logistic regression to obtain odds ratios (ORs) and 95% confidence intervals (CIs), or the chi-square test.
Results
Four SNPs in F5 (rs12120605, rs6427202, rs9332542 and rs6427199), one in F10 (rs3093261), and one in EPCR (rs2069948) were associated with breast cancer. EPCR rs2069948 was associated with estrogen receptor (ER) and progesterone receptor (PR) positivity, while the SNPs in F5 appeared to follow hormone receptor negative and triple negative patients. The prothrombotic polymorphisms factor V Leiden (rs6025) and prothrombin G20210A (rs1799963) were not associated with breast cancer. High APC resistance was associated with breast cancer in both factor V Leiden non-carriers (OR 6.5, 95% CI 4.1-10.4) and carriers (OR 38.3, 95% CI 6.2-236.6). The thrombin parameters short lag times (OR 5.8, 95% CI 3.7-9.2), short times to peak thrombin (OR 7.1, 95% CI 4.4-11.3), and high thrombin peak (OR 6.1, 95% CI 3.9-9.5) predicted presence of breast cancer, and high D-dimer also associated with breast cancer (OR 2.0, 95% CI 1.3-3.3). Among the coagulation inhibitors, low levels of antithrombin associated with breast cancer (OR 5.7, 95% CI 3.6-9.0). The increased coagulability was not explained by the breast cancer associated SNPs, and was unaffected by ER, PR and triple negative status.
Conclusions
A procoagulant phenotype was found in the breast cancer patients. Novel associations with SNPs in F5, F10 and EPCR to breast cancer susceptibility were demonstrated, and the SNPs in F5 were confined to hormone receptor negative and triple negative patients. The study supports the importance of developing new therapeutic strategies targeting coagulation processes in cancer.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2407-14-845) contains supplementary material, which is available to authorized users.
Keywords: Tissue factor pathway, Single nucleotide polymorphisms, Breast cancer, Activated protein C resistance, D-dimer, Genotype-phenotype correlations, Factor V Leiden, Prothrombin G20210A, Hormone receptor status, Triple negative status
Background
There is compelling evidence that blood coagulation and tumor biology are connected through multiple pathophysiological pathways. Disruption of the hemostatic balance is frequently observed in several cancer types [1–3]. The hypercoagulable state has been attributed to an adverse effect of malignant cells expressing procoagulants, but there is now evidence of a bidirectional interaction between cancer and coagulation [4]. Elevated plasma levels of activated coagulation markers, such as D-dimer, have been shown to be markers of cancer progression and poor outcome [5–7]. Plasma levels of the coagulation inhibitors antithrombin and protein C have been shown to decrease, while tissue factor (TF) pathway inhibitor (TFPI) was found to increase during cancer progression [8, 9], and several studies have shown that cancer patients acquire activated protein C (APC) resistance [2, 3, 10, 11].
During carcinogenesis a transcriptional program inducing expression of hemostatic genes is turned on [12]. Procoagulants produced by tumor- and stimulated host cells not only contribute to the increased risk of cancer-associated thrombosis, but can also trigger cancer- signaling pathways. Coagulation independent signaling increases the angiogenic and metastatic behavior of tumor cells and thereby accelerates the growth and spread of cancer. One of the most extensively studied procoagulants involved in cancer is TF. Induced by oncogenes, such as K-ras and the epidermal growth factor receptor (EGFR), TF is overexpressed in many cancers. TF expression has been shown to correlate with tumor progression and poor survival [13]. TF initiates the coagulation cascade by forming a complex with activated factor VII (FVIIa), which activates factor X (FX). The assembly of FXa with its activated cofactor, factor Va (FVa), leads to the generation of thrombin, fibrin formation and platelet activation. The activity of FXa and the TF/FVIIa catalytic complex is modulated by TFPI. In addition to coagulation activation, the TF complexes (TF/FVIIa and TF/FVIIa/FXa) may elicit G-protein coupled intracellular signaling mediated by protease- activated receptors (PARs). Activation of PAR-1 and PAR-2 results in expression of genes promoting angiogenesis, cell migration, proliferation, and metastasis [14]. Recently, it was demonstrated that the endothelial protein C receptor (EPCR) is able to bind the ternary TF/FVIIa/FXa complex and induce a more efficient PAR-1 and PAR-2 mediated signaling in endothelial cells [15]. EPCR positive breast cancer cells have an increased ability to form tumors in vivo [16].
The F5 rs6025 and F2 rs1799963 (commonly known as the factor V Leiden and the prothrombin G20210A polymorphisms, respectively) are well-established procoagulant polymorphisms that increase the risk of venous thrombosis, due to induction of APC resistance and increased levels of prothrombin, respectively. Mozsik et al. recently reported an association of factor V Leiden with gastrointestinal cancer [17], whereas Vossen et al. found a 6-fold increased risk of colorectal cancer for homozygous, but not for heterozygous factor V Leiden carriers [18]. Additional studies (across populations) on several cancer types have also failed to show an association with factor V Leiden heterozygotes [3, 19–22]. Except for Pihusch et al. [20], several studies have not been able to find an increased prevalence of the prothrombin G20210A polymorphism in cancer [3, 18, 19, 21, 22].
The F7 gene polymorphism -402GA (rs510317) has been reported to be associated with breast cancer [23]. Still, limited information on the role of polymorphisms in hemostatic genes to cancer pathogenesis is available, in particular regarding the more common variants.
Breast cancer is a highly heterogenous disease with substantial variation at both the clinical and the molecular level. Immunohistochemical expression of the growth regulating hormone receptors; estrogen receptor (ER) and progesterone receptor (PR), in addition to overexpression and/or amplification of the oncogene human epidermal growth factor receptor 2 (HER2) are clinically relevant markers for prognostic and predictive purposes. The majority of breast tumors (~80%) show hormone receptor positivity and are likely to respond to endocrine (hormonal) therapy. 10-15% of breast cancers belong to a subgroup called triple negative breast cancers, defined by lack of ER, PR and HER2 overexpression. Triple negative breast cancers tend to have poor prognosis, and currently, no targeted therapy has been approved for this type of breast cancer [24].
In the present case-control study, we aimed to investigate the role of common single nucleotide polymorphisms (SNPs) in genes involved in the TF pathway of coagulation (i.e., the F2, F3 (TF) F5, F7, F10, EPCR, and TFPI genes) on the susceptibility of breast cancer. In addition, markers of coagulation activity and plasma levels of coagulation inhibitors were measured, related to presence of breast cancer, and correlated to genotypes of breast cancer associated SNPs.
Methods
Patient material; cases and controls
The study comprised of 385 stage I or II female breast cancer patients (cases) enrolled between June 2008 and August 2010 at the Oslo University Hospital Ullevål, Oslo, and the Akershus University Hospital, Nordbyhagen, Norway. The cases were subjected to primary breast surgery (mastectomy or lumpectomy) without receiving any pre-operative treatment, and blood samples were drawn immediately before surgery. Cases that later were acknowledged to have metastatic disease were excluded. The controls comprised of 353 healthy women, who were originally recruited as controls in a study on the risk of venous thrombosis in pregnancy [25]. ER and PR status of the tumors were determined by immunohistochemistry and collected from pathology reviews, and tumor cell nuclei were scored according to pathology guidelines. HER2 status was determined by immunohistochemistry and/or by silver enhancement in situ hybridization (SISH) (Roche, Dual SISH HER-2) where a HER2 gene/centrosome 17 (CEP17) ratio of >2.2 defined HER2 positivity.
We excluded subjects who were not of Scandinavian descent (i.e., not from Norway, Sweden or Denmark) from genotypic and phenotypic analyzes, and subjects who were pregnant, or received anticoagulant- or hormone replacement therapy were excluded from the phenotypic analyzes. After excluding one case with metastases at the time of diagnosis, two cases that proved not to have breast cancer, and 16 non-Scandinavians, the final case group comprised of 366 breast cancer patients for both genotypic and phenotypic analyzes. Among the 353 control subjects, 46 were non-Scandinavian, thus leaving 307 controls for genotypic analyzes (i.e. SNPs). 34 controls were either pregnant or used oral contraceptives leaving 273 controls for phenotypic analyzes (i.e. hemostatic parameters). The average age at blood sampling was 57.7 (±11.2) (range 29-87) years for cases, and 40.0 (±5.6) (range 22-58) years for controls.
The Regional Committee for Medical and Health Research Ethics of South-East Norway approved the study (approval number 1.2006.1607, amendment 1.2007.1125 for Ullevål patients and 429-04148 for Akershus patients) and all included women gave their written informed consent to participate.
Blood sampling
Venous blood samples were collected in Vacutainer vacuum tubes (Becton-Dickinson, Plymouth, UK) containing 0.5 mL buffered sodium citrate (0.129 mol/L). Whole blood was centrifuged for 15 min at 2000 g at room temperature within 1 hour to prepare platelet poor plasma, and aliquots were stored at -70°C until analyzed. Using the same blood collection procedure as described, plasma from 21 healthy subjects (9 men and 12 women, mean age 43 years) were collected to create a pooled normal plasma (PNP) reference. None of these 21 subjects had antithrombin-, protein C- or protein S deficiencies, were carriers for the factor V Leiden or the prothrombin G20210A polymorphisms, tested positive for antiphospholipid antibodies (lupus anticoagulant, or anticardiolipin- or anti-β2-glycoprotein 1 antibodies), and they did not use oral contraceptives or any other hormones.
Phenotypic hemostatic parameters
The endogenous thrombin potential (ETP) was measured using the Calibrated Automated Thrombogram (CAT) assay [26], according to the manufacturer’s instructions (Thrombinoscope B.V, Maastricht, the Netherlands). Four thrombin generation parameters were recorded; ETP (time integral of the thrombin formation), lag time, peak thrombin (Peak), and time to peak thrombin (ttPeak). APC resistance was determined after the addition of APC (American Diagnostica Inc., Stamford, CT, USA), and the results were reported as APC-sensitivity ratio (APC-sr); which is the ratio of ETP in presence of APC divided by ETP in absence of APC normalized against the similar ratio obtained with PNP measured in the same run [25]. The coagulation inhibitors antithrombin and protein C activities, and free protein S antigen, were analyzed using the Chromogenix Coamatic® Antithrombin, the Chromogenix Coamatic® Protein C, and the Chromogenix Coamatic® Protein S-Free kits from Instrumentation Laboratory (Lexington, MA, USA). Free TFPI antigen and D-dimer were analyzed by the commercial enzyme-linked immunosorbent assay kits Asserachrom® Free TFPI and Asserachrom® D-DI from Diagnostica Stago, Asnières, France. The hemostatic parameters are shown in Additional file 1: Figure S1.
DNA isolation and genotyping
DNA was either isolated on the BioRobot Universal with the QIAamp DNA Blood BioRobot MDx Kit (Qiagen, Hilden, Germany) and eluted in Qiagen buffer AE (10 mM Tris-Cl 0.5 mM EDTA; pH 9.0), or with the Gentra Autopure LS machine using the Puregene Genomic DNA purification Kit (Gentra Systems, Minneapolis, MN 55441 USA), or manually using the MasterPure TM DNA Purification Kit for Blood Version II (Epicentre® Biotechnologies, Madison, WI, USA). SNPs were genotyped with the iPLEX Gold massarray platform (Sequenom) at the Centre for Integrative Genetics, Norwegian University of Life Sciences, Ås, Norway.
SNP selection and quality control
We used a SNP tagging approach to avoid genotyping redundant SNPs. By using a minor allele frequency (MAF) criterion of ≥10% and pairwise r2 ≥ 0.8 as a cut-off for proxies, 39 SNPs were selected in the following gene regions: F2 (n = 3), F3 (TF) (n = 4), F5 (n = 10), F7 (n = 2), F10 (n = 9), TFPI (n = 9), and EPCR (n = 2). The tag-SNP selection was performed using the Tagger program ( http://www.broad.mit.edu/mpg/haploview/, [27]) implemented in Haploview v. 4.2 and genotype data from the Caucasian population (Utah residents with ancestry from northern and western Europe) from the HapMap project release 27, phase III on NCBI B36 assembly, dbSNPb126. Factor V Leiden (rs6025) and the prothrombin G20210A (rs1799963) polymorphisms were also included in the SNP selection. Hence, the final SNP selection consisted of 41 SNPs that were genotyped in both cases and controls.
Individuals with ≥50% missing genotypes and SNPs with <97% call rates were excluded for further analysis. SNPs that deviated from Hardy-Weinberg equilibrium in the exact test were also excluded (significance threshold P < 0.001).
One SNP failed genotyping in all individuals (TFPI rs3213739), two SNPs had genotyping call rates <97% (F7 rs1475931 and TFPI rs2041778), and one SNP was out of Hardy Weinberg equilibrium in controls (EPCR rs867186). Hence, after filtering, 37 of the 41 genotyped SNPs remained for further analysis.
Statistical methods
All statistical analyzes were performed using SPSS statistical software (version 21.0; SPSS Inc., Chicago, IL, USA) and PLINK v.1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/).
Associations between each SNP and breast cancer were analyzed using an allelic chi-square (χ2) test with 1 degree of freedom. The false discovery rate (FDR) procedure described by Benjamini & Hochberg [28] was used to correct for multiple testing.
Odds ratios (ORs), 95% confidence intervals (CIs), and P-values were determined for the genotypes of the SNPs that were significant at the 5% level and had a FDR < 0.25 in the allelic test. Binary logistic regression under the additive risk model was applied with case/control status as the dependent variable, and genotypes (coded 0, 1, 2 for each extra risk allele) as the categorical independent variables. Risk alleles were defined as the alleles being more prevalent among cases, thus ORs >1 were obtained.
Independence between SNP associations was tested by conditional analysis in PLINK, where the allelic dosage for a given SNP was added as a covariate in a binary logistic regression model (additive model). The E-M algorithm was used to estimate haplotype frequencies, and haplotype-based association analysis was conducted using binary logistic regression (additive model). Haploview v. 4.2 was used for creating linkage disequilibrium (LD) plots, and the SNAP tool [29] was used to obtain pair-wise LD measurements. The Alamut software (v. 2.0) was used to predict if any of the associated SNPs, or their proxies, could affect splicing.
The plasma levels of hemostatic parameters were compared between cases and controls using t-test when normally distributed, or the non-parametric Mann-Whitney when the distribution was skewed. Tests with P < 0.05 were considered significantly different. Logistic regression was used to determine the associations with breast cancer status, or ER, PR, HR, and triple negative status, for either high or low levels of each of the hemostatic parameters. Case and control subjects were dichotomized according to either the 10th or the 90th percentiles of the hemostatic parameters’ plasma levels (defined in controls). The group with levels above the 10th percentile or below the 90th percentile served as the reference group.
Genotype-phenotype correlations were evaluated by the Kruskal-Wallis test. For tests with P < 0.05, follow-up pairwise comparisons were conducted using Mann Whitney U testing with Bonferroni correction, and genotype-phenotype pairs with at least one significant pairwise test were reported. For correlations with the APC resistant phenotype, factor V Leiden carriers were excluded due to the established role for the factor V Leiden variant in APC resistance.
Results
Associations between SNPs in TF pathway genes and risk of breast cancer
Associations between SNP alleles of the TF pathway genes and risk of breast cancer were assessed by comparing allele distributions between cases and controls (Table 1). A total of six SNPs in three distinct genes exhibited significantly different allele distributions with FDR < 0.25 (four in F5: rs12120605 (P = 0.026), rs6427202 (P = 0.028), rs9332542 (P = 0.023), rs6427199 (P = 0.037), one in F10: rs3093261 (P = 0.011), and the one in EPCR: rs2069948 (P = 0.030)).
Table 1.
Allele distributions of SNPs in TF pathway genes in cases and controls
| Gene | Chr | SNP | Bp (hg18) | Region | Minor allele | Major allele | Freqency cases | Frequency controls | χ 2 | OR | P-value (unadj.) | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F2 | 11 | rs2070852 | 46701501 | Intronic | C | G | 0.290 | 0.316 | 0.99 | 0.89 | 0.319 | 0.83 |
| F2 | 11 | rs3136516 | 46717332 | Intronic | G | A | 0.448 | 0.448 | 0.00 | 1.00 | 0.996 | 1.00 |
| F2 | 11 | rs5896 | 46701579 | Coding (T > M) | T | C | 0.133 | 0.127 | 0.09 | 1.05 | 0.760 | 0.93 |
| F2 | 11 | rs1799963 | 46717631 | 3UTR | A | G | 0.013 | 0.005 | 2.20 | 2.60 | 0.138 | 0.61 |
| F3 | 1 | rs3917643 | 94774455 | Intronic | C | T | 0.058 | 0.080 | 2.58 | 0.70 | 0.109 | 0.57 |
| F3 | 1 | rs696619 | 94777808 | Intronic | A | G | 0.414 | 0.440 | 0.87 | 0.90 | 0.351 | 0.83 |
| F3 | 1 | rs1324214 | 94769876 | Intronic | A | G | 0.240 | 0.271 | 1.59 | 0.85 | 0.207 | 0.70 |
| F3 | 1 | rs3917615 | 94774578 | Intronic | T | C | 0.439 | 0.433 | 0.04 | 1.02 | 0.839 | 0.93 |
| F5 | 1 | rs12120605 | 167789178 | Intronic | T* | G | 0.140 | 0.100 | 4.93 | 1.47 | 0.026 | 0.22 |
| F5 | 1 | rs6427202 | 167795454 | Intronic | C* | T | 0.454 | 0.394 | 4.82 | 1.28 | 0.028 | 0.22 |
| F5 | 1 | rs9332542 | 167805907 | Intronic | A | G* | 0.286 | 0.344 | 5.16 | 0.76 | 0.023 | 0.22 |
| F5 | 1 | rs6427199 | 167790161 | Intronic | A | G* | 0.364 | 0.420 | 4.36 | 0.79 | 0.037 | 0.23 |
| F5 | 1 | rs6012 | 167795204 | Intronic | T | C | 0.162 | 0.164 | 0.01 | 0.98 | 0.906 | 0.93 |
| F5 | 1 | rs4524 | 167778379 | Coding (K > R) | C | T | 0.261 | 0.257 | 0.03 | 1.02 | 0.871 | 0.93 |
| F5 | 1 | rs4656687 | 167771782 | Intronic | C | T | 0.323 | 0.312 | 0.17 | 1.05 | 0.676 | 0.93 |
| F5 | 1 | rs6025 | 167785673 | Coding (Q > R) | T | C | 0.033 | 0.034 | 0.02 | 0.96 | 0.900 | 0.93 |
| F5 | 1 | rs9287095 | 167805090 | Intronic | A | G | 0.094 | 0.104 | 0.34 | 0.90 | 0.561 | 0.93 |
| F5 | 1 | rs10158595 | 167786988 | Intronic | T | C | 0.223 | 0.248 | 1.14 | 0.87 | 0.286 | 0.83 |
| F5 | 1 | rs9332618 | 167767105 | Intronic | A | G | 0.133 | 0.139 | 0.08 | 0.95 | 0.773 | 0.93 |
| F7 | 13 | rs491098 | 112817347 | Intronic | C | G | 0.106 | 0.117 | 0.39 | 0.90 | 0.530 | 0.93 |
| F10 | 13 | rs3093261 | 112824083 | Near 5UTR | T* | C | 0.460 | 0.391 | 6.41 | 1.33 | 0.011 | 0.22 |
| F10 | 13 | rs3211744 | 112832999 | Intronic | T | G | 0.136 | 0.154 | 0.85 | 0.87 | 0.358 | 0.83 |
| F10 | 13 | rs2026160 | 112840894 | Intronic | C | A | 0.284 | 0.258 | 1.08 | 1.14 | 0.298 | 0.83 |
| F10 | 13 | rs9549675 | 112846885 | Intronic | T | C | 0.205 | 0.239 | 2.10 | 0.83 | 0.148 | 0.61 |
| F10 | 13 | rs3211719 | 112825510 | Intronic | G | A | 0.252 | 0.245 | 0.09 | 1.04 | 0.769 | 0.93 |
| F10 | 13 | rs3211752 | 112835460 | Intronic | G | A | 0.499 | 0.489 | 0.13 | 1.04 | 0.716 | 0.93 |
| F10 | 13 | rs556694 | 112828042 | Intronic | C | T | 0.093 | 0.089 | 0.04 | 1.04 | 0.836 | 0.93 |
| F10 | 13 | rs3211770 | 112841850 | Intronic | A | G | 0.109 | 0.113 | 0.05 | 0.96 | 0.818 | 0.93 |
| F10 | 13 | rs473598 | 112849190 | Intronic | A | G | 0.128 | 0.137 | 0.23 | 0.92 | 0.629 | 0.93 |
| EPCR | 20 | rs2069948 | 33226150 | Intronic | C* | T | 0.468 | 0.408 | 4.72 | 1.27 | 0.030 | 0.22 |
| TFPI | 2 | rs2192825 | 188099064 | Intronic | C | T | 0.439 | 0.402 | 1.81 | 1.16 | 0.178 | 0.66 |
| TFPI | 2 | rs7594359 | 188117093 | Intronic | T | C | 0.461 | 0.441 | 0.52 | 1.08 | 0.472 | 0.93 |
| TFPI | 2 | rs2192824 | 188077036 | Intronic | T | C | 0.449 | 0.428 | 0.54 | 1.09 | 0.461 | 0.93 |
| TFPI | 2 | rs13424790 | 188032097 | Downstream | G | T | 0.317 | 0.310 | 0.08 | 1.03 | 0.778 | 0.93 |
| TFPI | 2 | rs8176548 | 188048580 | Intronic | T | C | 0.351 | 0.347 | 0.02 | 1.02 | 0.878 | 0.93 |
| TFPI | 2 | rs10187622 | 188122406 | Intronic | T | C | 0.161 | 0.165 | 0.03 | 0.97 | 0.864 | 0.93 |
| TFPI | 2 | rs12613071 | 188096556 | Intronic | C | T | 0.202 | 0.195 | 0.09 | 1.04 | 0.762 | 0.93 |
P-values were determined by the χ2 -test. Alleles for the positive DNA strand (UCSC annotated) are shown. Significantly associated SNPs; bold, and factor V Leiden (F5 rs6025) and prothrombin G20210A (F2 rs1799963); italic. *Risk alleles for significant SNPs.
Chr: chromosome. OR: Odds ratio as determined for the minor allele with the major allele as reference. FDR: False discovery rate as described by Benjamini and Hochberg [28].
The genotype distributions for these six SNPs were compared using an additive model, since the underlying genetic model was unknown. All loci showed significant associations also at the genotypic level, with ORs ranging from 1.27 to 1.49 (Additional file 2: Table S1).
Furthermore, whereas the association with EPCR rs2069948 was restricted to patients with ER and PR positive tumors (OR 1.27, 95% CI 1.01-1.58; and OR 1.30, 95% CI 1.03-1.65, respectively), there was an overall trend that the four SNPs in F5 were confined to hormone receptor negative patients (ER/PR negative) (ORs from 1.54 to 1.99) and triple negative patients (ER/PR/HER2 negative) (ORs from 1.68 to 2.11) when compared with healthy controls (Additional file 3: Table S2). However, the analyzes lacked power to show significant differences in genotype distributions between patient subgroups.
The allele distribution for factor V Leiden was equal among cases (3.3%) and controls (3.4%), while the prothrombin G20210A polymorphism appeared more frequently (non-significant) in patients (1.3%) compared to controls (0.5%) (Table 1). Only heterozygous carriers of either polymorphism were detected.
According to the SNAP tool, each of the F5 rs6427202, F5 rs9332542, and EPCR rs2069948 SNPs were all in strong or perfect LD (r2 = 0.93-1.00) with several SNPs in the European (CEU) population. The F5 rs6427202 SNP was in strong LD (r2 ≥ 0.96) with 20 intronic F5 SNPs, but also with five SNPs in the P-selectin coding gene; SELP (r2 ≥ 0.93). Moreover, in the Regulome database (RegulomeDB) [30], the F5 rs9332542 and four SNPs in perfect LD (rs2227245, rs2213872, rs2213873, rs6662176) were annotated as cis-acting expression quantitative trait loci (eQTL) for F5, and the F10 rs3093261 was predicted to be an eQTL for the LAMP1 gene encoding lysosome-associated membrane protein 1 (LAMP-1), located ~175 kb downstream of F10 rs3093261. Using the Alamut software, none of the SNPs within a 300 bp distance from the nearest splice site, appeared to affect splicing.
Conditional- and haplotype analysis of the F5SNPs associated with breast cancer
In total, four breast cancer associated SNPs were found in the F5 gene region (rs12120605, rs6427202, rs9332542, and rs6427199), and the interdependence of these SNPs on breast cancer risk was investigated (Table 2). The rs12120605 appeared to represent an independent signal, as this SNP remained significantly associated after conditioning on the other three F5 SNPs as separate covariates. Moreover, when set as the conditional SNP, the rs12120605 did not diminish the significance of the other three SNPs, reflecting the modest pairwise LD with these SNPs (D’ ≤ 0.50) (Figure 1). In contrast, a dependency appeared to exist between the rs6427202, rs9332542, and rs6427199 as their effects were neutralized when conditioned on each other. This result, combined with the LD structure between the three SNPs (D’ between 0.35-1.00) (Figure 1), pointed towards a potential haplotype effect. Indeed, the haplotype consisting of all three individual risk alleles (C-G-G) was common in the population (frequency of 0.32), and was found to be significantly associated with breast cancer (OR 1.39, P = 0.011). Conditioning on factor V Leiden did not alter the association of any of the other F5 SNPs (data not shown).
Table 2.
Results of the conditional association analysis for the four significant F5 gene SNPs
| SNP | Original ORs (P-values) | ORs (P-values) conditioned on rs12120605 | ORs (P-values) conditioned on rs6427202 | ORs (P-values) conditioned on rs9332542 | ORs (P-values) conditioned on rs6427199 |
|---|---|---|---|---|---|
| rs12120605 | 1.49 (0.024) | - | 1.55 (0.015) | 1.54 (0.016) | 1.62 (0.009) |
| rs6427202 | 1.32 (0.021) | 1.36 (0.010) | - | 1.19 (0.218) | 1.24 (0.085) |
| rs9332542 | 1.32 (0.022) | 1.35 (0.014) | 1.20 (0.244) | - | 1.23 (0.095) |
| rs6427199 | 1.27 (0.036) | 1.33 (0.014) | 1.22 (0.111) | 1.20 (0.141) | - |
ORs and P-values are shown before and after conditioning on each of the SNPs. Significant conditional associations are shown in italic.
Figure 1.
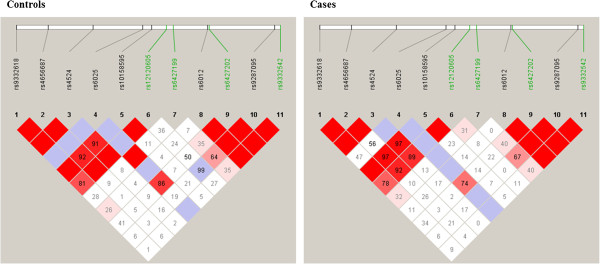
Linkage disequilibrium (LD) plots of the F5 SNPs. Linkage disequilibrium (LD) plots of the 11 analyzed SNPs including factor V Leiden (rs6025), within the F5 gene for controls (left plot) and cases (right plot). The LD measure D’ is shown. The disease associated SNPs are depicted in green.
Relation between hemostatic parameters and breast cancer
Coagulation activity and levels of coagulation inhibitors were compared between cases and controls to explore if any hemostatic abnormalities existed. Median levels and P-values are provided in Additional file 4: Table S3.
The estimated associations between breast cancer status and either high or low levels of the hemostatic parameters are shown in Table 3. From the CAT assay, an association with breast cancer was predicted for lag times and times to peak thrombin below the 10th percentile, and for peak thrombin above the 90th percentile, with ORs ranging from 5.8 to 7.1. ETP above the 90th percentile did not associate with breast cancer. APC resistance levels above the 90th percentile associated with breast cancer in factor V Leiden non-carriers (OR 6.5, 95% CI 4.06-10.35), but also in factor V Leiden carriers (OR 38.3, 95% CI 6.2-236.6). Moreover, subjects with high D-dimer levels (>90th percentile) were also associated with breast cancer disease (OR 2.0, 95% CI 1.26-3.28). No association with breast cancer was found for low levels of the coagulation inhibitors protein C, protein S or free TFPI, but for antithrombin activity below the 10th percentile, the association with breast cancer was ~6-fold higher compared to activity above the 10th percentile. None of the associated hemostatic parameters were specific to the different subsets of patients, as defined by the ER and PR hormone receptor status or triple negative status (Additional file 5: Table S4).
Table 3.
Distribution of cases and controls among the high or low level categories of the hemostatic parameters
| Hemostatic parameter | Cases (n) | Controls (n) | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| Coagulation activity: | |||||
| ETP (%)* | |||||
| <90th percentile | 309 | 246 | Ref. | Ref. | Ref. |
| >90th percentile | 37 | 27 | 1.09 | 0.65-1.84 | 0.744 |
| Lag time (%)* | |||||
| >10th percentile | 211 | 246 | Ref. | Ref. | Ref. |
| <10th percentile | 135 | 27 | 5.83 | 3.71-9.16 | <0.001 |
| ttPeak (%)* | |||||
| >10th percentile | 206 | 249 | Ref. | Ref. | Ref. |
| <10th percentile | 140 | 24 | 7.05 | 4.40-11.29 | <0.001 |
| Peak (%)* | |||||
| <90th percentile | 208 | 246 | Ref. | Ref. | Ref. |
| >90thpercentile | 138 | 27 | 6.05 | 3.85-9.50 | <0.001 |
| APC resistance (nAPC-sr) | |||||
| FV Leiden non-carriers | |||||
| <90th percentile | 190 | 228 | Ref. | Ref. | Ref. |
| >90th percentile | 135 | 25 | 6.48 | 4.06-10.35 | <0.001 |
| FV Leiden carriers | |||||
| <90th percentile | 4 | 18 | Ref. | Ref. | Ref. |
| >90th percentile | 17 | 2 | 38.3 | 6.2-236.6 | <0.001 |
| D-dimer (ng/mL) | |||||
| <90th percentile | 292 | 246 | Ref. | Ref. | Ref. |
| >90th percentile | 65 | 27 | 2.03 | 1.26-3.28 | 0.004 |
| Coagulation inhibitors: | |||||
| AT (%) | |||||
| >10th percentile | 215 | 246 | Ref. | Ref. | Ref. |
| <10th percentile | 135 | 27 | 5.72 | 3.64-8.99 | <0.001 |
| Protein C (%) | |||||
| >10th percentile | 310 | 246 | Ref. | Ref. | Ref. |
| <10th percentile | 40 | 27 | 1.18 | 0.702-1.97 | 0.539 |
| Protein S (%) | |||||
| >10th percentile | 325 | 246 | Ref. | Ref. | Ref. |
| <10th percentile | 22 | 27 | 0.62 | 0.34-1.11 | 0.106 |
| Free TFPI (ng/mL) | |||||
| >10th percentile | 319 | 246 | Ref. | Ref. | Ref. |
| <10th percentile | 39 | 27 | 1.11 | 0.66-1.87 | 0.683 |
ORs, 95% CI and P-values were obtained by logistic regression with respect to cases.
AT = antithrombin. ttpETP = time to thrombin peak. nAPC-sr = normalised APC sensitivity ratio. (%) describes activity as compared to pooled normal plasma (PNP). *CAT-assay variables.
Since increased APC resistance in cancer has been detected in several previous studies, adjustments were made for the hemostatic parameters that correlated to APC resistance; protein C (ρ = -0.18, P = 0.003), protein S (ρ = -0.33, P < 0.001), and free TFPI (ρ = -0.42, P < 0.001). Adjusting for each of these parameters as separate covariates had only a modest impact on the association between high APC resistance and breast cancer in factor V Leiden non-carriers (data not shown), and the OR obtained in the full model with all covariates included (OR 8.6, 95% CI 5.1-14.3), was similar to that of the unadjusted model in Table 3 (OR 6.5, 95% CI 4.06-10.35). Equivalent results were obtained for factor V Leiden carriers (adjusted OR 50.7, 95% CI 6.6-390.9 vs. unadjusted OR 38.3, 95% CI 6.2-236.6 (Table 3)). Age did not correlate to any of the associated coagulation parameters (assessed in controls), except for an inverse correlation to APC resistance (ρ = -0.13, P = 0.032).
Genotype-phenotype associations
In an effort to explain some of the hemostatic alterations observed in the patients, we searched for possible regulatory relationships between the genetic variations and the hemostatic parameters. Only the hemostatic parameters being significantly altered were considered, and we explored if any of these parameters were unevenly distributed across the genotypes of the six breast cancer associated SNPs in Table 1 (and Additional file 2: Table S1). Genotype-phenotype associations were made separately for controls and cases since divergent regulatory mechanisms could exist in the two groups. Two significant correlations were found in the control group; F5 rs6427202 correlated with thrombin peak (Figure 2A), whereas F5 rs6427199 correlated with antithrombin (Figure 2B). Because high thrombin peak and low antithrombin activity were found associated with breast cancer (Table 3), we adjusted for F5 rs6427202 and F5 rs6427199, respectively. However, these adjustments did not change the OR estimates obtained in the unadjusted model (data not shown). Interestingly, a trend for a correlation between the number of F5 rs6427199 risk alleles and high APC resistance was found in controls (P = 0.15). F5 rs6427199 may thus be an interesting candidate for general investigations of novel APC resistance inducing factors. No genotype-phenotype correlations were found in the case group.
Figure 2.
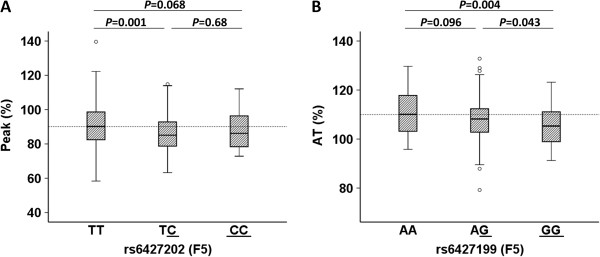
Significant genotype-phenotype correlations. Significant genotype-phenotype correlations in the control group after pairwise comparison analyzed by the Mann Whitney U test. Tests with ≥1 significant Bonferroni corrected P-value(s) are shown. (Bonferroni corrected P; 0.05/9 = 0.006). Distribution of (A) Peak (%) across the genotypes of F5 rs6427202, and (B) antithrombin (%) across the genotypes of F5 rs6427199. The box and whiskers plots show the minimum, 25th percentile, median, 75th percentile and maximum levels of the hemostatic parameters plotted against the genotype of the SNPs. The risk alleles of the SNPs are underlined.
Discussion
Hypercoagulability is a common, but complex and multifactorial phenomenon in cancer. Although involvement of both clinical and biological aspects is recognized, the precise mechanism(s) underlying how the hemostatic system relates to cancer is not clear.
Among the 37 common SNPs in seven TF pathway genes (TF, F2, F5, F7, F10, TFPI and EPCR), six SNPs in three separate genes were found to be associated with breast cancer: four intronic SNPs in the F5 gene (rs12120605, rs6427202, rs9332542 and rs6427199), one in the upstream region of the F10 gene (rs3093261), and one intronic SNP in the EPCR gene (rs2069948). This is first-time evidence for an association between these SNPs and cancer.
Since breast cancer is a heterogeneous disease, certain risk factors may be specific for subsets of patients. In this study, the SNP in EPCR was associated with patients positive for ER and PR, while the SNPs in F5 showed a tendency towards a preferential association with hormone receptor negative patients and triple negative patients. This might indicate that the F5 SNPs may influence breast cancer etiology in hormone receptor negative/triple negative patients.
The association with F5 expression [31] represents a possible link to the increased coagulation activity in breast cancer, and F10 rs3093261, located between the F10 and F7 gene, has been associated with increased FVII levels in stroke patients [32], which may link F10 rs3093261 to the increased coagulation activation observed in our study.
Supporting a role in cancer, EPCR has shown tumor growth promoting effects in mice [16].
Notably, the associated F5 SNPs were common variants, and were independent of factor V Leiden carrier status. We found no association with factor V Leiden heterozygosity. Although not previously investigated in untreated breast cancer, studies in gastric- [19], gynaecological- [21], colorectal- [3], and oral cancer [22, 33] support this lack of association. So far, the study by Vossen et al. is the only study large enough to investigate the significance of homozygous factor V Leiden, reporting a 6-fold increased risk of colorectal cancer in homozygous carriers [18]. Interestingly, a reduced cancer risk (~30%) for heterozygous factor V Leiden carriers was reported in the same study [18]. Neither did we find a significant association with the prothrombin G20210A variant. However, given the low allele frequency (0.5-1.3%), a larger sample size would be needed to establish the role of this variant in breast cancer risk. Correspondingly, other studies across cancer types did not succeed in finding an association with prothrombin G20210A [3, 19, 21, 22, 33]. Pihusch et al. found an increased risk of gastrointestinal cancer for this variant [20], while Vossen et al. found a decreased risk of colorectal cancer for prothrombin G20210A heterozygotes [18].
In addition to the SNP discoveries, our study also provides evidence of a hypercoagulable state in the breast cancer patients, as detected by the CAT assay and increased APC resistance and D-dimer. The CAT assay parameters lag time, time to peak thrombin and peak thrombin were all associated with breast cancer, but not total ETP. Hence, the combination of the kinetic CAT parameters is likely to provide a better understanding of hypercoagulable states than do individual parameters [34]. Although care should be taken in comparing different thrombin assays [35], our CAT assay results are in line with previous studies in breast cancer reporting increased levels of thrombin formation [2], and shortened activated partial thromboplastin times (aPTT) [36].
We also found that increased APC resistance was associated with breast cancer. This finding is supported by a former breast cancer study [2], and studies in colorectal- [3] and gastrointestinal cancer [10]. In line with the existing literature, levels of protein S, TFPI, and also protein C correlated inversely to APC resistance [2, 37]. However, these potential APC resistance determinants did not affect the association between APC resistance and breast cancer. Interestingly, we found that increased APC resistance was associated with breast cancer in both carriers and non-carriers of factor V Leiden. Supported by previous studies [1, 38], this suggests that the acquired APC resistant phenotype appears more crucial in cancer than APC resistance caused by factor V Leiden. Acquired APC resistance could be due to increased levels of coagulation factors like factor V and factor VIII [11], yet unidentified factors produced by tumor- or stimulated host cells, or novel hereditary causes. Of note, the factor V Cambridge (rs118203906) variant, previously associated with increased APC resistance [39], turned out to be monomorphic in our case subjects (data not shown).
Further demonstrating the procoagulant state, high levels of the fibrin degradation product D-dimer were also associated with breast cancer. This confirms previous studies in breast cancer [6, 36, 40], and also other cancers like colorectal- [41], lung- [42] and gastric cancer [43]. These studies also demonstrated D-dimer as a potential important marker in disease stage prediction and for prognostic purposes. Furthermore, increased levels of D-dimer in cancer have been detected even in the absence of thrombosis [44].
Among the coagulation inhibitors, only low levels of antithrombin were associated with breast cancer. Decreased antithrombin levels have previously been found in breast cancer by Nijziel et al. [2], while no difference was observed either in another breast cancer study [45], a study on colorectal- [3] or a study on advanced cancers [1]. Supporting our observations, the two latter studies also found that neither protein S nor protein C levels in patients deviated from healthy controls. In a study of gastrointestinal cancer, Lindahl et al. found that the activity of antithrombin and protein C decreased [8], while the TFPI activity increased as the cancer progressed. Later, Iversen et al. confirmed that the median levels of TFPI activity were above the upper normal limit in gastrointestinal- and lung cancer, and in metastatic patients. In contrast, median TFPI activity in breast cancer was within the normal range [9]. Comparable to the latter, we found no difference in levels of TFPI in the cases compared to controls. Thus, the TFPI levels may vary according to cancer type and disease stage.
Noteworthy, the hypercoagulability did not seem to depend on hormone receptor status, as the estimated associations between breast cancer and the hemostatic plasma markers were not significantly different between hormone receptor negative or triple negative patients and patients with other subtypes. This observation is in agreement with a recent breast cancer study that did not observe any variation in D-dimer, prothrombin times (PT), and aPTT according to ER or PR status [36].
Since breast cancer is among the cancers with the lowest risk of thrombosis [46], our study indicates that activated coagulation may reflect the biology of the underlying tumor and could be an important indicator of cancer progression. In this context, it should be emphasized that the herein retrospective data is only suited to assess the presence of a hypercoagulable state in patients already diagnosed with breast cancer. Prospective studies are needed to establish whether coagulation activation precedes breast tumor development, or if it mirrors the course of the established disease. Interestingly, a prospective study reported a ~3-fold increased rate of digestive tract cancers in men with persistent coagulation activation [47].
The present work represents the most comprehensive study to investigate hemostasis in a homogenous (Scandinavian) breast cancer material. The tag SNP selection ensures good coverage of the normal genetic diversity within each selected gene. In this respect, it should be noted that our SNP associations may be a reflection of LD with other known or yet unknown true causal variants. In order to verify the significance of the associated SNPs, our findings should be validated in a different study population and other cancer types.
One limitation of the study is that the breast cancer patients were older than the control women, thus, an age-related bias could exist for the comparison of hemostatic plasma markers between patients and controls. Although supported by the existing literature, these results should therefore be interpreted with some caution. On the other hand, the inclusion criteria eliminate a possible effect of anticoagulant- or hormone therapy, and pregnancy, as well as chemotherapy, on the hemostatic parameters.
Conclusions
This study has established the existence of a global procoagulant profile in breast cancer patients. The coagulation activity seemed to be independent of factor V Leiden and prothrombin G20210A. Instead, novel associations between common SNPs in genes of the TF pathway (F5, F10, and EPCR) and breast cancer susceptibility were demonstrated. Based on both phenotypic and genotypic evidence, this study supports the importance of developing new therapeutic strategies targeting coagulation processes in cancer.
Electronic supplementary material
Additional file 1: Figure S1: TF-pathway of coagulation. The TF-FVIIa complex initiates the coagulation cascade by activating FX to FXa, which aided by its cofactor FVa cleaves prothrombin to generate thrombin. Thrombin cleaves fibrinogen to form fibrin monomers that together with activated platelets form a blood clot. D-dimer is a fibrin degradation product. Coagulation inhibitors and their targets are designated. EPCR has been indicated to associate with the TF-FVIIa complex. The FVL (rs6025) polymorphism prevents the ability of aPC to cleave and inhibit FVa, causing (inherited) APC resistance. The PT G20210A (rs1799963) polymorphism increases the rate of prothrombin protein production. Regular arrows and blunt-end arrows illustrate activation and inhibition, respectively. The hemostatic markers measured in plasma in this study are shaded in grey. EPCR = endothelial protein C receptor, TF = tissue factor, FVIIa = activated factor VII, FX = factor X, FXa = activated factor X, FVa = activated factor Va, TFPI = tissue factor pathway inhibitor, PS = protein S, aPC = activated protein C, AT = antithrombin, PT = prothrombin, FVL = factor V Leiden, APCR = activated protein C resistance. (PDF 160 KB)
Additional file 2: Table S1: Genotype distributions of the significant SNPs in TF pathway genes in cases and controls. ORs, 95% CI and P-values determined with respect to the risk allele (bold) using logistic regression (additive model). Alleles for the positive DNA strand (UCSC annotated) are shown. (PDF 100 KB)
Additional file 3: Table S2: The breast cancer associated SNPs stratified by hormone receptor status (ER and PR) and triple negative status (ER negative/PR negative/HER2 negative) (additive model in binary logistic regression). ORs determined with respect to the risk allele. Significant associations are shown in bold. (PDF 188 KB)
Additional file 4: Table S3: Plasma levels of hemostatic parameters in cases and controls. Median values with IQR shown in brackets. (PDF 90 KB)
Additional file 5: Table S4: ORs and P-values for hemostatic parameters stratified by hormone receptor status (ER and PR) and triple negative status (ER negative/PR negative/HER2 negative). (PDF 99 KB)
Acknowledgement
We would like to thank Oslo Breast Cancer Consortium (OSBREAC) for providing patient samples, and Marie-Christine Mowinckel and Brit Steinsvik for conducting the plasma protein measurements. The South-Eastern Norway Regional Health Authority, Hamar, Norway, the Norwegian Radium Hospital Research Foundation, and A. Jahre and H.G Andrine Berg & son foundations provided research grants.
Abbreviations
- TF
Tissue factor
- APC
Activated protein C
- TFPI
Tissue factor pathway inhibitor
- ER
Estrogen receptor
- PR
Progesterone receptor
- HR
Hormone receptor
- OR
Odds ratio
- EPCR
Endothelial protein C receptor
- HER2
Human epidermal growth factor receptor 2
- SNP
Single nucleotide polymorphism
- ETP
Endogenous thrombin potential
- CAT
Calibrated automated thrombogram
- ttPeak
Time to Peak
- MAF
Minor allele frequency
- FDR
False discovery rate
- CI
Confidence interval
- LD
Linkage disequilibrium.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MT, MKV, AED, GS, BAL PMS and NI participated in the design of the study. NI conceived the study. MT, MKV, AED, GS, BAL, PMS and NI interpreted the data. MT performed the statistical analysis and drafted the manuscript. MKV participated in the statistical analysis. AED, HKMV, KKS, ØG, ALBD, AFJ, VK, IB, RK, ES, PMS, NI participated in acquisition of the data. MKV, AED, HKMV, KKS, ØG, ALBD, AFJ, VK, IB, RK, ES, GS, BAL, PMS, NI critically revised the intellectual content. All authors read and approved the final manuscript.
Contributor Information
Mari Tinholt, Email: mari.tinholt@medisin.uio.no.
Marte Kathrine Viken, Email: m.k.viken@medisin.uio.no.
Anders Erik Dahm, Email: aeadahm@gmail.com.
Hans Kristian Moen Vollan, Email: hans.kristian.vollan@me.com.
Kristine Kleivi Sahlberg, Email: kristine.kleivi@gmail.com.
Øystein Garred, Email: UXYSGA@ous-hf.no.
Anne-Lise Børresen-Dale, Email: a.l.borresen-dale@medisin.uio.no.
Anne Flem Jacobsen, Email: UXAFJA@ous-hf.no.
Vessela Kristensen, Email: v.n.kristensen@medisin.uio.no.
Ida Bukholm, Email: i.r.k.bukholm@medisin.uio.no.
Rolf Kåresen, Email: rolf.karesen@medisin.uio.no.
Ellen Schlichting, Email: ELSCHL@ous-hf.no.
Grethe Skretting, Email: grethe.skretting@medisin.uio.no.
Benedicte Alexandra Lie, Email: b.a.lie@medisin.uio.no.
Per Morten Sandset, Email: p.m.sandset@medisin.uio.no.
Nina Iversen, Email: nina.iversen@medisin.uio.no.
References
- 1.Green D, Maliekel K, Sushko E, Akhtar R, Soff GA. Activated-protein-C resistance in cancer patients. Haemostasis. 1997;27:112–118. doi: 10.1159/000217442. [DOI] [PubMed] [Google Scholar]
- 2.Nijziel MR, van Oerle R, Christella M, Thomassen LG, van Pampus EC, Hamulyak K, Tans G, Rosing J. Acquired resistance to activated protein C in breast cancer patients. Br J Haematol. 2003;120:117–122. doi: 10.1046/j.1365-2141.2003.03987.x. [DOI] [PubMed] [Google Scholar]
- 3.Paspatis GA, Sfyridaki A, Papanikolaou N, Triantafyllou K, Livadiotaki A, Kapsoritakis A, Lydataki N. Resistance to activated protein C, factor V leiden and the prothrombin G20210A variant in patients with colorectal cancer. Pathophysiol Haemost Thromb. 2002;32:2–7. doi: 10.1159/000057282. [DOI] [PubMed] [Google Scholar]
- 4.Franchini M, Montagnana M, Favaloro EJ, Lippi G. The bidirectional relationship of cancer and hemostasis and the potential role of anticoagulant therapy in moderating thrombosis and cancer spread. Semin Thromb Hemost. 2009;35:644–653. doi: 10.1055/s-0029-1242718. [DOI] [PubMed] [Google Scholar]
- 5.Kohli M, Fink LM, Spencer HJ, Zent CS. Advanced prostate cancer activates coagulation: a controlled study of activation markers of coagulation in ambulatory patients with localized and advanced prostate cancer. Blood Coagul Fibrinolysis. 2002;13:1–5. doi: 10.1097/00001721-200201000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Dirix LY, Salgado R, Weytjens R, Colpaert C, Benoy I, Huget P, van Dam P, Prove A, Lemmens J, Vermeulen P. Plasma fibrin D-dimer levels correlate with tumour volume, progression rate and survival in patients with metastatic breast cancer. Br J Cancer. 2002;86:389–395. doi: 10.1038/sj.bjc.6600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ay C, Dunkler D, Pirker R, Thaler J, Quehenberger P, Wagner O, Zielinski C, Pabinger I. High D-dimer levels are associated with poor prognosis in cancer patients. Haematologica. 2012;97:1158–1164. doi: 10.3324/haematol.2011.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl AK, Sandset PM, Abildgaard U, Andersson TR, Harbitz TB. High plasma levels of extrinsic pathway inhibitor and low levels of other coagulation inhibitors in advanced cancer. Acta Chir Scand. 1989;155:389–393. [PubMed] [Google Scholar]
- 9.Iversen N, Lindahl AK, Abildgaard U. Elevated TFPI in malignant disease: relation to cancer type and hypercoagulation. Br J Haematol. 1998;102:889–895. doi: 10.1046/j.1365-2141.1998.00875.x. [DOI] [PubMed] [Google Scholar]
- 10.De Lucia D, De Vita F, Orditura M, Renis V, Belli A, Conte M, di Grazia M, Iacoviello L, Donati MB, Catalano G. Hypercoagulable state in patients with advanced gastrointestinal cancer: evidence for an acquired resistance to activated protein C. Tumori. 1997;83:948–952. doi: 10.1177/030089169708300615. [DOI] [PubMed] [Google Scholar]
- 11.Sarig G, Michaeli Y, Lanir N, Brenner B, Haim N. Mechanisms for acquired activated protein C resistance in cancer patients. J Thromb Haemost. 2005;3:589–590. doi: 10.1111/j.1538-7836.2005.01168.x. [DOI] [PubMed] [Google Scholar]
- 12.Boccaccio C, Comoglio PM. Genetic link between cancer and thrombosis. J Clin Oncol. 2009;27:4827–4833. doi: 10.1200/JCO.2009.22.7199. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg YW, Osanto S, Reitsma PH, Versteeg HH. The relationship between tissue factor and cancer progression: insights from bench and bedside. Blood. 2012;119:924–932. doi: 10.1182/blood-2011-06-317685. [DOI] [PubMed] [Google Scholar]
- 14.Schaffner F, Ruf W. Tissue factor and protease-activated receptor signaling in cancer. Semin Thromb Hemost. 2008;34:147–153. doi: 10.1055/s-2008-1079254. [DOI] [PubMed] [Google Scholar]
- 15.Disse J, Petersen HH, Larsen KS, Persson E, Esmon N, Esmon CT, Teyton L, Petersen LC, Ruf W. The endothelial protein C receptor supports tissue factor ternary coagulation initiation complex signaling through protease-activated receptors. J Biol Chem. 2011;286:5756–5767. doi: 10.1074/jbc.M110.201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaffner F, Yokota N, Carneiro-Lobo T, Kitano M, Schaffer M, Anderson GM, Mueller BM, Esmon CT, Ruf W. Endothelial protein C receptor function in murine and human breast cancer development. PLoS One. 2013;8:e61071. doi: 10.1371/journal.pone.0061071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mozsik G, Rumi G, Domotor A, Figler M, Gasztonyi B, Papp E, Par A, Par G, Belagyi J, Matus Z, Melegh B. Involvement of serum retinoids and Leiden mutation in patients with esophageal, gastric, liver, pancreatic, and colorectal cancers in Hungary. World J Gastroenterol. 2005;11:7646–7650. doi: 10.3748/wjg.v11.i48.7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vossen CY, Hoffmeister M, Chang-Claude JC, Rosendaal FR, Brenner H. Clotting factor gene polymorphisms and colorectal cancer risk. J Clin Oncol. 2011;29:1722–1727. doi: 10.1200/JCO.2010.31.8873. [DOI] [PubMed] [Google Scholar]
- 19.Battistelli S, Stefanoni M, Genovese A, Vittoria A, Cappelli R, Roviello F. Prevalence of factor V Leiden and prothrombin G20210A in patients with gastric cancer. World J Gastroenterol. 2006;12:4179–4180. doi: 10.3748/wjg.v12.i26.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pihusch R, Danzl G, Scholz M, Harich D, Pihusch M, Lohse P, Hiller E. Impact of thrombophilic gene mutations on thrombosis risk in patients with gastrointestinal carcinoma. Cancer. 2002;94:3120–3126. doi: 10.1002/cncr.10590. [DOI] [PubMed] [Google Scholar]
- 21.Tormene D, Beltramello P, Perlati M, Brandolin B, Barbar S, De Toffoli G, Simioni P. The risk of cancer progression in women with gynecological malignancies and thrombophilic polymorphisms: a pilot case-control study. Clin Appl Thromb Hemost. 2009;15:535–539. doi: 10.1177/1076029608317941. [DOI] [PubMed] [Google Scholar]
- 22.Vairaktaris E, Yapijakis C, Wiltfang J, Ries J, Vylliotis A, Derka S, Vasiliou S, Neukam FW. Are factor V and prothrombin mutations associated with increased risk of oral cancer? Anticancer Res. 2005;25:2561–2565. [PubMed] [Google Scholar]
- 23.Eroglu A, Ozturk A, Akar N. Association between the -402GA, -401GT, and -323ins10-bp polymorphisms of factor VII gene and breast cancer. Breast Cancer. 2011;18:282–285. doi: 10.1007/s12282-009-0189-6. [DOI] [PubMed] [Google Scholar]
- 24.Vuong D, Simpson PT, Green B, Cummings MC, Lakhani SR. Molecular classification of breast cancer. Virchows Arch. 2014;465:1–14. doi: 10.1007/s00428-014-1593-7. [DOI] [PubMed] [Google Scholar]
- 25.Bergrem A, Dahm AE, Jacobsen AF, Mowinckel MC, Sandvik L, Sandset PM. Resistance to activated protein C is a risk factor for pregnancy-related venous thrombosis in the absence of the F5 rs6025 (factor V Leiden) polymorphism. Br J Haematol. 2011;154:241–247. doi: 10.1111/j.1365-2141.2011.08712.x. [DOI] [PubMed] [Google Scholar]
- 26.Hemker HC, Giesen P, Al Dieri R, Regnault V, de Smedt E, Wagenvoord R, Lecompte T, Beguin S. Calibrated automated thrombin generation measurement in clotting plasma. Pathophysiol Haemost Thromb. 2003;33:4–15. doi: 10.1159/000071636. [DOI] [PubMed] [Google Scholar]
- 27.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 29.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, Schnabel RB, Lubos E, Mennerich D, Rust W, Perret C, Proust C, Nicaud V, Loscalzo J, Hubner N, Tregouet D, Munzel T, Ziegler A, Tiret L, Blankenberg S, Cambien F. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zakai NA, Lange L, Longstreth WT, Jr, O’Meara ES, Kelley JL, Fornage M, Nikerson D, Cushman M, Reiner AP. Association of coagulation-related and inflammation-related genes and factor VIIc levels with stroke: the Cardiovascular Health Study. J Thromb Haemost. 2011;9:267–274. doi: 10.1111/j.1538-7836.2010.04149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vylliotis A, Yapijakis C, Nkenke E, Nisyrios T, Avgoustidis D, Adamopoulou M, Ragos V, Vassiliou S, Koronellos N, Vairaktaris E. Effect of thrombosis-related gene polymorphisms upon oral cancer: a regression analysis. Anticancer Res. 2013;33:4033–4039. [PubMed] [Google Scholar]
- 34.Tripodi A, Legnani C, Palareti G, Chantarangkul V, Mannucci PM. More on: high thrombin generation and the risk of recurrent venous thromboembolism. J Thromb Haemost. 2009;7:906–907. doi: 10.1111/j.1538-7836.2009.03338.x. [DOI] [PubMed] [Google Scholar]
- 35.Stepien E, Plicner D, Branicka A, Stankiewicz E, Pazdan A, Sniezek-Maciejewska M, Gorkiewicz I, Kapelak B, Sadowski J. Factors influencing thrombin generation measured as thrombin-antithrombin complexes levels and using calibrated automated thrombogram in patients with advanced coronary artery disease. Pol Arch Med Wewn. 2007;117:297–305. [PubMed] [Google Scholar]
- 36.Tas F, Kilic L, Duranyildiz D. Coagulation tests show significant differences in patients with breast cancer. Tumour Biol. 2014;35:5985–5992. doi: 10.1007/s13277-014-1793-4. [DOI] [PubMed] [Google Scholar]
- 37.Negaard HF, Iversen PO, Ostenstad B, Mowinckel MC, Sandset PM. Increased acquired activated protein C resistance in unselected patients with hematological malignancies. J Thromb Haemost. 2008;6:1482–1487. doi: 10.1111/j.1538-7836.2008.03060.x. [DOI] [PubMed] [Google Scholar]
- 38.Zangari M, Saghafifar F, Anaissie E, Badros A, Desikan R, Fassas A, Mehta P, Morris C, Toor A, Whitfield D, Siegel E, Barlogie B, Fink L, Tricot G. Activated protein C resistance in the absence of factor V Leiden mutation is a common finding in multiple myeloma and is associated with an increased risk of thrombotic complications. Blood Coagul Fibrinolysis. 2002;13:187–192. doi: 10.1097/00001721-200204000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Williamson D, Brown K, Luddington R, Baglin C, Baglin T. Factor V Cambridge: a new mutation (Arg306 → Thr) associated with resistance to activated protein C. Blood. 1998;91:1140–1144. [PubMed] [Google Scholar]
- 40.Blackwell K, Haroon Z, Broadwater G, Berry D, Harris L, Iglehart JD, Dewhirst M, Greenberg C. Plasma D-dimer levels in operable breast cancer patients correlate with clinical stage and axillary lymph node status. J Clin Oncol. 2000;18:600–608. doi: 10.1200/JCO.2000.18.3.600. [DOI] [PubMed] [Google Scholar]
- 41.Oya M, Akiyama Y, Okuyama T, Ishikawa H. High preoperative plasma D-dimer level is associated with advanced tumor stage and short survival after curative resection in patients with colorectal cancer. Jpn J Clin Oncol. 2001;31:388–394. doi: 10.1093/jjco/hye075. [DOI] [PubMed] [Google Scholar]
- 42.Gabazza EC, Taguchi O, Yamakami T, Machishi M, Ibata H, Suzuki S. Evaluating prethrombotic state in lung cancer using molecular markers. Chest. 1993;103:196–200. doi: 10.1378/chest.103.1.196. [DOI] [PubMed] [Google Scholar]
- 43.Tas F, Ciftci R, Kilic L, Serilmez M, Karabulut S, Duranyildiz D. Clinical and prognostic significance of coagulation assays in gastric cancer. J Gastrointest Cancer. 2013;44:285–292. doi: 10.1007/s12029-013-9490-x. [DOI] [PubMed] [Google Scholar]
- 44.Knowlson L, Bacchu S, Paneesha S, McManus A, Randall K, Rose P. Elevated D-dimers are also a marker of underlying malignancy and increased mortality in the absence of venous thromboembolism. J Clin Pathol. 2010;63:818–822. doi: 10.1136/jcp.2010.076349. [DOI] [PubMed] [Google Scholar]
- 45.Yigit E, Gonullu G, Yucel I, Turgut M, Erdem D, Cakar B. Relation between hemostatic parameters and prognostic/predictive factors in breast cancer. Eur J Intern Med. 2008;19:602–607. doi: 10.1016/j.ejim.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 46.Chew HK, Wun T, Harvey DJ, Zhou H, White RH. Incidence of venous thromboembolism and the impact on survival in breast cancer patients. J Clin Oncol. 2007;25:70–76. doi: 10.1200/JCO.2006.07.4393. [DOI] [PubMed] [Google Scholar]
- 47.Miller GJ, Bauer KA, Howarth DJ, Cooper JA, Humphries SE, Rosenberg RD. Increased incidence of neoplasia of the digestive tract in men with persistent activation of the coagulant pathway. J Thromb Haemost. 2004;2:2107–2114. doi: 10.1111/j.1538-7836.2004.01011.x. [DOI] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here: http://www.biomedcentral.com/1471-2407/14/845/prepub
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1: TF-pathway of coagulation. The TF-FVIIa complex initiates the coagulation cascade by activating FX to FXa, which aided by its cofactor FVa cleaves prothrombin to generate thrombin. Thrombin cleaves fibrinogen to form fibrin monomers that together with activated platelets form a blood clot. D-dimer is a fibrin degradation product. Coagulation inhibitors and their targets are designated. EPCR has been indicated to associate with the TF-FVIIa complex. The FVL (rs6025) polymorphism prevents the ability of aPC to cleave and inhibit FVa, causing (inherited) APC resistance. The PT G20210A (rs1799963) polymorphism increases the rate of prothrombin protein production. Regular arrows and blunt-end arrows illustrate activation and inhibition, respectively. The hemostatic markers measured in plasma in this study are shaded in grey. EPCR = endothelial protein C receptor, TF = tissue factor, FVIIa = activated factor VII, FX = factor X, FXa = activated factor X, FVa = activated factor Va, TFPI = tissue factor pathway inhibitor, PS = protein S, aPC = activated protein C, AT = antithrombin, PT = prothrombin, FVL = factor V Leiden, APCR = activated protein C resistance. (PDF 160 KB)
Additional file 2: Table S1: Genotype distributions of the significant SNPs in TF pathway genes in cases and controls. ORs, 95% CI and P-values determined with respect to the risk allele (bold) using logistic regression (additive model). Alleles for the positive DNA strand (UCSC annotated) are shown. (PDF 100 KB)
Additional file 3: Table S2: The breast cancer associated SNPs stratified by hormone receptor status (ER and PR) and triple negative status (ER negative/PR negative/HER2 negative) (additive model in binary logistic regression). ORs determined with respect to the risk allele. Significant associations are shown in bold. (PDF 188 KB)
Additional file 4: Table S3: Plasma levels of hemostatic parameters in cases and controls. Median values with IQR shown in brackets. (PDF 90 KB)
Additional file 5: Table S4: ORs and P-values for hemostatic parameters stratified by hormone receptor status (ER and PR) and triple negative status (ER negative/PR negative/HER2 negative). (PDF 99 KB)


