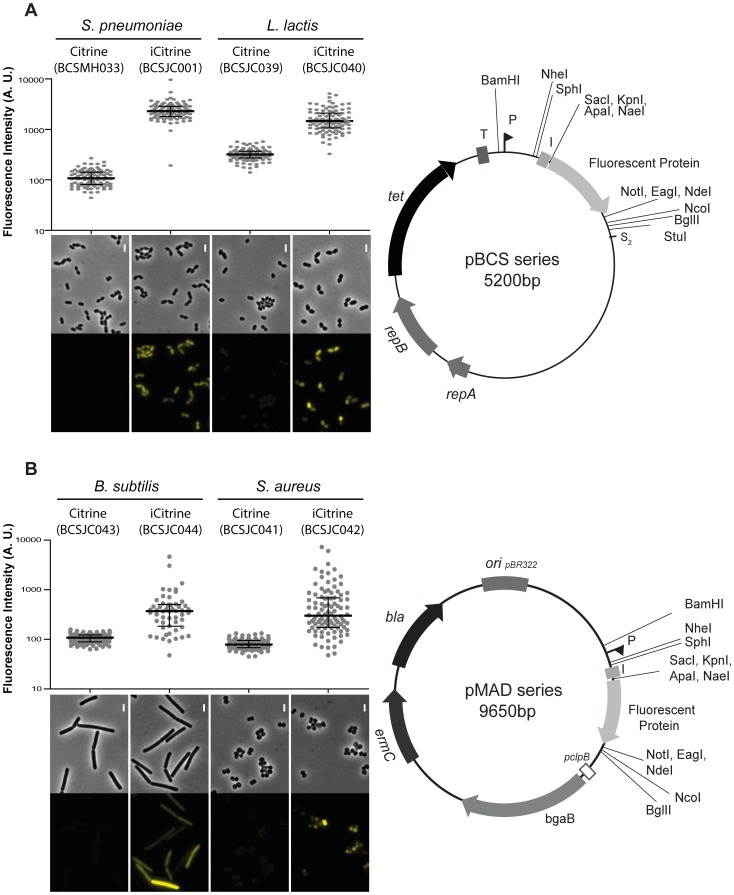
Figure 1. Linking the i-tag to Citrine improves the expression of fluorescence in Gram-positive bacteria.
A) (Left panel) Median fluorescence, with 25% and 75% inter-quartile range (black lines) of the fluorescence signal detected in S. pneumoniae unencapsulated bacteria, in arbitrary units (A. U.), expressing Citrine (strain BCSMH033) or iCitrine (strain BCSJC001), and L. lactis, expressing Citrine (strain BCSJC039) and iCitrine (strain BCSJC040). At least 100 cells of each strain were quantified. Representative images of each strain are shown below the graph. Scale bar: 1 µm. (Right panel) Map of the pBCS plasmids. Fluorescent protein refers to Citrine and iCitrine. repA, repB, plasmid replication genes. tet, tetracycline resistance marker. T, transcription terminator. P, promoter. S2, stop codon. B) (Left panel) Median fluorescence, with 25% and 75% inter-quartile range (black lines) of the fluorescence signal emitted by Citrine and iCitrine detected in S. aureus bacteria, in arbitrary units (A. U.), expressing Citrine (strain BCSJC041) and iCitrine (strain BCSJC042), or B. subtilis, expressing Citrine (strain BCSJC043) or iCitrine (strain BCSJC044). At least 100 cells of each strain were quantified. Representative images of each strain are shown below the graph. Scale bar: 1 µm. (Right panel) Map of the pMAD plasmids. Fluorescent protein refers to Citrine and iCitrine. Unique restriction sites are indicated. ermC, erythromycin resistance marker.