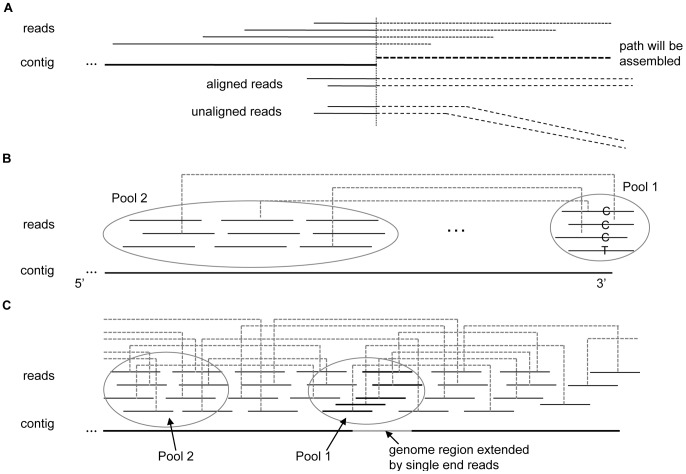
Figure 2. Align reads to contig for extension.
(A) Align reads to contig. The path of nearby genome region is extracted according to the reads that are partially aligned onto contig, then new reads having more than 90% aligned bases with the path will be considered correctly aligned; otherwise, they should be aligned onto other genome regions rather than at that position. (B) Extension using paired-end reads. Contig is extended at the 3′ end according to the reads in Pool 1 and whose mates in Pool 2. There are two candidate bases ‘C’ and ‘T’, and ‘C’ is well supported by the mates in the two pools, whereas ‘T’ has no paired-end reads support, thus ‘C’ will be chosen to append onto the contig. (C) Extension using single-end reads. When assemble the grey color region which cannot be assembled by paired-end reads and the reads in Pool 1 have no mates in Pool 2, the reads in Pool 1 are used as single-end reads to extend the contig.