Abstract
Corynebacterium jeikeium is an opportunistic pathogen which has been noted for significant genomic diversity. The population structure within this species remains poorly understood. Here we explore the relationships among fifteen clinical isolates of C. jeikeium (reference strains K411 and ATCC 43734, and 13 primary isolates collected over a period of 7 years) through genetic, genomic, and phenotypic studies. We report a high degree of divergence among strains based on 16S ribosomal RNA (rRNA) gene and rpoB gene sequence analysis, supporting the presence of genetically distinct subgroups. Whole genome sequencing indicates genomic-level dissimilarity among subgroups, which qualify as 4 separate and distinct Corynebacterium species based on an Average Nucleotide Identity (ANIb) threshold of < 95%. Functional distinctions in antibiotic susceptibilities and metabolic profiles characterize two of these genomospecies, allowing their differentiation from others through routine laboratory testing. The remaining genomospecies can be classified through a biphasic approach integrating phenotypic testing and rpoB gene sequencing. The genomospecies predominantly recovered from patient specimens does not include either of the existing C. jeikeium reference strains, implying that studies of this pathogen would benefit from examination of representatives from the primary disease-causing group. The clinically dominant genomospecies also has the smallest genome size and gene repertoire, suggesting the possibility of increased virulence relative to the other genomospecies. The ability to classify isolates to one of the four C. jeikeium genomospecies in a clinical context provides diagnostic information for tailoring antimicrobial therapy and may aid in identification of species-specific disease associations.
Keywords: genomics, speciation, phylogeny, clinical testing, classification, Corynebacterium jeikeium
Introduction
Corynebacterium jeikeium is a clinically important opportunistic pathogen capable of causing a wide range of disorders including endocarditis, septicemia, joint infection, pneumonia, osteomyelitis, meningitis, and soft tissue infection (Cazanave et al., 2012; Funke et al., 1997; Ifantidou et al., 2010; Tleyjeh et al., 2005), particularly in immunocompromised patients or those with indwelling medical devices. It is recognized as the most frequently recovered medically significant Corynebacterium species among patients in intensive care facilities, with the capacity for nosocomial dissemination (Funke et al., 1997; Tauch et al., 2005).
Previous work (Riegel et al., 1994) has sought to investigate the range of genomic, physiological, and phenotypic differences displayed by this organism. Although DNA-DNA hybridization (DDH) studies revealed considerable genomic diversity among C. jeikeium isolates, biochemical testing was unable to further delineate groups among C. jeikeium strains (Riegel et al., 1994). All isolates in that study were consequently assigned to a single species under one of four “genomic groups”. Nevertheless, it was noted that some groups displayed differences in their antibiotic susceptibilities (Riegel et al., 1994), hinting at dissimilarities in underlying physiology. To the best of our knowledge, the genomic diversity and population structure of C. jeikeium has not been revisited in the 20 years subsequent to that publication.
Recently, whole-genome sequencing technologies have made it possible to more comprehensively explore the genomic content and population structure of bacteria (Chan et al., 2012; Georgiades and Raoult, 2010), allowing for the robust classification of prokaryotes into meaningful taxonomic groups based on discrete and quantitative metrics (Richter and Roselló- Móra, 2009). Such approaches would be beneficial in the exploration of the evolutionary relationships among C. jeikeium strains, however, at this time only the complete genome of C. jeikeium reference strain K411(Tauch et al., 2005) and an incomplete genome of ATCC type strain 43734 (Jackman et al., 1987; Peterson et al., 2009) are available for such analyses. To better understand the population structure and diversity of C. jeikeium strains, here we have performed whole genome sequencing of 13 C. jeikeium primary clinical isolates. We use genomic and phenotypic data to explore the relationships among available strains and to revisit the current classification of C. jekeium in light of genomic-era techniques (Richter and Roselló- Móra, 2009).
Materials and Methods
Isolates
C. jeikeium strains were isolated from patients in our hospital or other hospitals in the US Pacific Northwest (Table 1), representing all C. jeikeium isolates sent to our laboratory for diagnostic molecular identification from the years 2006 to 2012. Reference strain C. jeikeium K411 (Kerry-Williams and Noble, 1984; Tauch et al., 2005) was obtained from the National Collection of Type Cultures (London, United Kingdom). C. jeikeium ATCC type strain 43734 (Jackman et al., 1987) was obtained from the American Type Culture Collection (Manassas, Virginia, United States). All strains were cultured aerobically at 37°C on sheep blood agar plates. DNA was extracted from isolates using Ultraclean Microbial DNA Isolation kit (MoBio).
Table 1.
Primary Clinical Isolates and Assembly Statistics
| Isolate name | Source | Year Isolated |
Bases Assembled |
N50 Statistic (bp) |
Number of Contigs |
Estimated d% GC |
Estimated Fold Coverage |
Genbank Accession for Draft Genome |
Genbank Accession for partial 16S rRNA sequence |
Genbank Accession for partial rpoB sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| Cj47447 | Blood (Catheter) | 2006 | 2421042 | 72298 | 139 | 61.45 | 154 | JFCM00000000 | KJ526268 | KJ526290 |
| Cj14566 | Unknown | 2009 | 2336582 | 119061 | 71 | 61.89 | 98 | JFCF00000000 | KJ526269 | KJ526282 |
| Cj16348 | Unknown | 2009 | 2335869 | 142412 | 68 | 61.71 | 113 | JFCG00000000 | KJ526270 | KJ526288 |
| Cj19409 | Spine | 2010 | 2361515 | 133002 | 72 | 61.8 | 105 | JFCI00000000 | KJ526271 | KJ526281 |
| Cj21382 | Humeral membrane | 2010 | 2281623 | 93369 | 165 | 61.9 | 31 | JFCJ00000000 | KJ526272 | KJ526283 |
| Cj47445 | Blood (Catheter) | 2011 | 2513217 | 101031 | 142 | 61.33 | 125 | JFCO00000000 | KJ526280 | KJ526291 |
| Cj30184 | Unknown | 2011 | 2431357 | 55484 | 401 | 61.85 | 55 | JFCK00000000 | KJ526278 | KJ526284 |
| Cj30952 | Vertebral disk space | 2011 | 2258819 | 134848 | 58 | 62.95 | 64 | JFCR00000000 | KJ526274 | KJ526289 |
| Cj47446 | Decubitus ulcer | 2011 | 2386733 | 80571 | 121 | 61.63 | 116 | JFCN00000000 | KJ526273 | KJ526293 |
| Cj37130 | Knee bursa | 2012 | 2304265 | 257322 | 41 | 61.93 | 124 | JFCQ00000000 | KJ526275 | KJ526285 |
| Cj38002 | Unknown | 2012 | 2310143 | 170201 | 59 | 62.03 | 121 | JFCP00000000 | KJ526279 | KJ526286 |
| Cj47453 | Pleural fluid | 2012 | 2393249 | 175261 | 94 | 61.58 | 84 | JFCL00000000 | KJ526276 | KJ526287 |
| Cj47444 | Pleural fluid | 2012 | 2430056 | 100676 | 97 | 62.33 | 131 | JFCH00000000 | KJ526277 | KJ526292 |
| C. jeikeium ATCC 43734 | Blood (Endocarditis) | 1987 | 2426461* | 26091 | 93 | 61.6* | N/A# | ACYW01000000 | CJU87823 | ACYW01000000* |
| C. jeikeium K411 | Axilla | 1984 | 2462499* | N/A | N/A | 61.40* | N/A | CR931997 | CR931997* | CR931997* |
from existing genome assembly
not applicable
16S rRNA and rpoB gene sequencing
Taxonomically informative 16S rRNA and rpoB gene fragments were PCR amplified from bacterial genomic DNA and sequenced using the Sanger method to establish gene sequences for 16S rRNA variable regions V1 to V3 (first ~500bp) and a fragment of RNA polymerase subunit gene, rpoB as described elsewhere (Khamis et al., 2005; Pottumarthy et al., 2003), or analogous sequences were extracted from published sequence data (Table 1).
Whole genome sequencing
100 ng of each genomic DNA was digested for 2 hours at 37°C in a 10 µl volume using 0.3 µl NEBNext dsDNA Fragmentase (New England Biolabs). DNA was simultaneously end-repaired and A-tailed in a 40 µl reaction containing 1× Rapid Ligation Buffer (Enzymatics Inc.), 0.1675 mM each dNTP (New England Biolabs), 0.1 µl E. coli DNA Polymerase I (New England Biolabs), 0.5 µl T4 PNK (New England Biolabs), and 0.02 µl Taq DNA Polymerase (New England Biolabs), incubated at 37°C for 30 minutes and 72°C for 20 minutes. Annealed Y-adaptors (5’-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’ and 5’-P GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG-3’, P = phosphorylation) were added at a concentration of 0.2 µM and ligated at 25°C for 20 minutes using T4 DNA Ligase in Rapid Ligation Buffer (Enzymatics). After purification with AmPure beads (Agencourt), the library was PCR amplified with KAPA HiFi HotStart ReadyMix using primer PRECAP_FWD_AMP_COMMON (5’- AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGC-3’) and sample-specific barcoded primers (5’- CAAGCAGAAGACGGCATACGAGATXXXXXXXXCGGTCTCGGCATTCCTGCTGAACCG- 3’), where X’s indicate the position of an 8 bp sample-specific index. Cycling conditions were (95°C for 3 min, 10 cycles of 98°C for 20 seconds, 65°C for 15 seconds, and 72°C for 1 minute, followed by one cycle of 72°C for 5 minutes). PCR product was purified with AmPure beads, pooled, and sequenced on a MiSeq (Illumina) using 250 bp Paired-End Reads with a custom index primer (5’-AGATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCG-3’). All oligonucleotides were synthesized by IDT.
Data Analysis
Adaptors were trimmed using the program Fastq-Mcf (http://code.google.com/p/ea-utils/) with skew filtering disabled and other parameters at their default. Draft genomes were assembled using AbySS v1.3.5 (Simpson et al., 2009). The N50 statistic (the length of which half of all contigs are equal to or larger) for assemblies was calculated using AbySS. Average Nucleotide Identity by BLAST (ANIb) values were calculated for draft genomes using jSpecies v1.2.1 (Richter and Roselló-Móra, 2009). Variants were called by aligning short read data from sequenced strains and published sequence read data from C. jeikeium ATCC 43734 (SRA ID numbers SRX037287 and SRX 002393) against C. jekeium K411 (GenBank accession no CR931997.1) using BWA version 0.6.1-r104 (Li and Durbin, 2009) and SAMtools version 0.1.18 (Li et al., 2009). Single nucleotide variant and indel calling was next performed using VarScan version 2.3.6 (Koboldt et al., 2012) with a minimum read depth (defined as the number of sequence reads covering a variant site) of 5 and a minimum variant frequency (defined as the fraction of reads contain a given variant) of 0.75. Estimated read depth per strain was estimated by the Lander-Waterman method (Lander and Waterman, 1988). Neighbor-Joining phylogenetic trees for single gene targets and whole genome sequence variants were generated by SplitsTree4 (Huson and Bryant, 2006), using 1,000 bootstrap replicates to estimate reliability. Comparative genomic analyses were visualized using BRIG version 0.95 (Alikhan et al., 2011) with default parameters. Gene prediction and annotation for assemblies was performed using the RAST server version 4.0 (Aziz et al., 2008) with default parameters.
Mass spectrometry, biochemical characterization, and antibiotic susceptibility profiling
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Spectrometric classification of strains was performed using the MALDI Biotyper system and Biotypes software (Bruker Daltonics). Biochemical testing of isolates was performed using the RapID™ CB Plus system (Remel). Antibiotic susceptibility testing was carried out using the Etest® method (Biomerieux).
Sequence data availability
Sanger sequence from 16S rRNA and rpoB gene fragments, and draft genome assemblies for all isolates are publically available through GenBank (Table 1). Sequence data generated for this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under study accession number SRP045192.
Results
Phylogenetic analysis of 16S rRNA and rpoB gene sequences
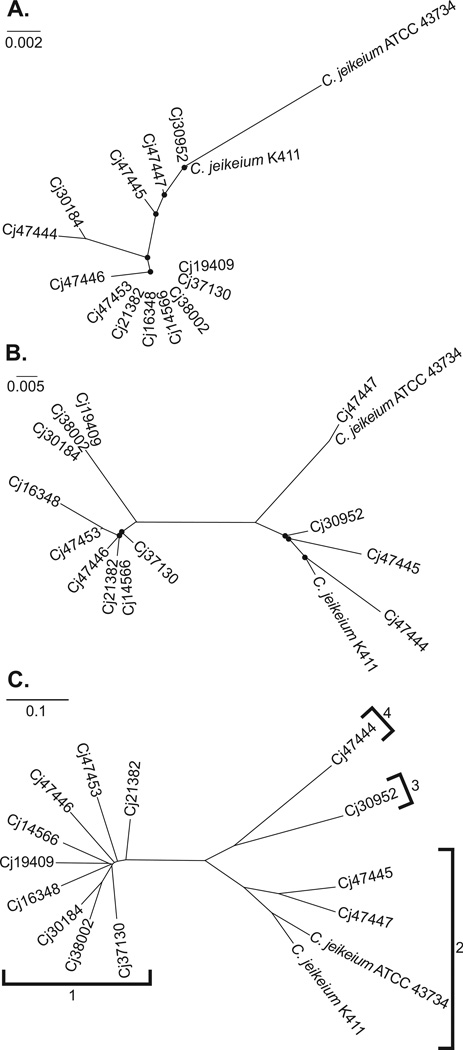
We examined all C. jeikeium isolates identified by our laboratory over a period of seven years, which should therefore comprise a relatively unbiased and representative sampling of the taxon as it is encountered clinically. Prior to initiation of these studies, the classification of all isolates as C. jeikeium was confirmed to high confidence (score ≥ 2.0) by mass spectrometry. We initially examined the strain collection through targeted sequencing of taxonomically informative genes, comparing our results to those of the fully sequenced reference strain K411 (Tauch et al., 2005) and partially sequenced reference ATCC type strain 43734 (Jackman et al., 1987). The 16S rRNA gene is a useful target for classifying bacterial species (Clarridge, 2004), and phylogenetic analysis of partial 16S rRNA gene sequence data from our isolates suggested the presence of genetically distinct C. jeikeium subdivisions (Figure 1A), with one clade encompassing most of the clinical isolates. Nevertheless, 16S rRNA gene sequence data can sometimes prove misleading or unreliable in inferring bacterial population structure (Georgiades and Raoult, 2010), especially among Corynebacterium species, where 16S rRNA gene diversity tends to be relatively low (Khamis et al., 2005). As an alternative approach, we therefore sequenced rpoB, a housekeeping gene that has proven informative for the molecular identification and classification of Corynebacterium isolates (Khamis et al., 2005). Phylogenetic analysis of rpoB also revealed substantial population structure suggesting the presence of genetically distinct groups (Figure 1B), although placement of strains to particular clades within the larger trees was not fully consistent when comparing 16S rRNA and rpoB gene targets.
Figure 1. Genetic and genomic phylogenies of primary C. jeikeium isolates and reference strains K411 and ATCC 43734.
A. Phylogeny constructed from partial 16S rRNA gene sequence (regions V1 – V2). B. Phylogeny constructed from partial rpoB gene sequence. C. Phylogeny constructed from whole genome variation. Brackets indicate distinct genomospecies as defined in Table 2. Scale bars for each phylogeny are indicated below the corresponding letter code, and are expressed in “changes per site”. Nodes overlaid with a black dot indicate a bootstrap value of < 75%.
Whole genome sequencing and identification of genomospecies
To more thoroughly explore the genomic differences displayed among the collected strains, we next performed whole genome sequencing and de novo genome assembly of each clinical isolate. Sequencing was performed to an average read depth of 100 × (and no less than an average of 30 × per sample) with respect to the C. jeikeium reference genome. For all isolates considered, the estimated GC content and number of nucleotides assembled for clinical isolates were similar to reported values for C. jeikeium K411 and ATCC 43734 (Table 1).
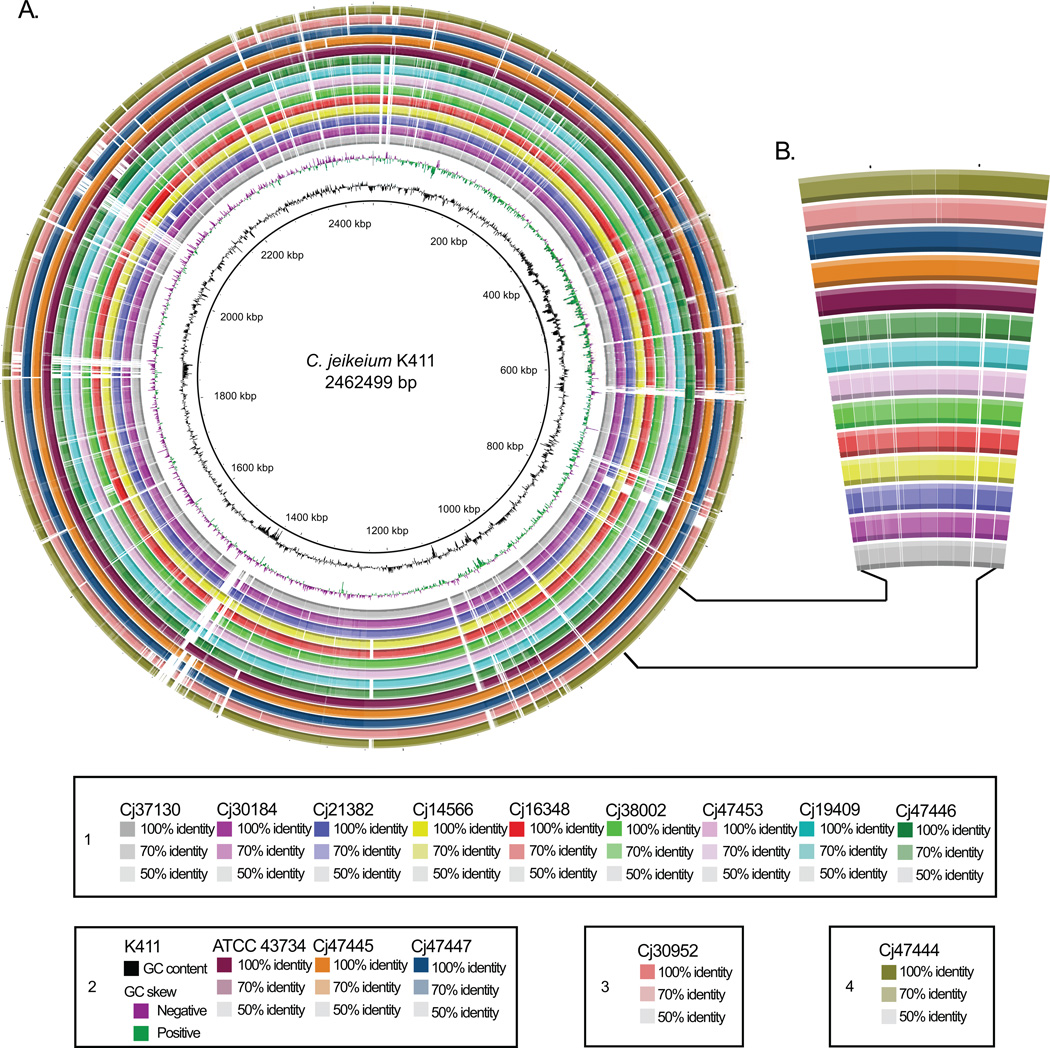
A phylogeny based on variants (single nucleotide polymorphisms and indels) identified across the whole genomes of strains independently supported division of isolates among distinct taxa (Figure 1C). Both broad partitions, as well as finer-scale relationships within the partitions, were all supported by high confidence (> 75%) bootstrap values. In the genomic phylogeny, reference strains K411 and ATCC 43734 formed a clade along with two primary isolates (Cj47447 and Cj47445). Two other strains (Cj30952 and Cj47444) were related to this group, but together constituted a separate clade. The remaining 9 isolates, representing the majority of strains examined, formed a distinct group more distantly related to other clades. The structure of the tree deduced from whole-genome variation closely resembled that of the rpoB-based tree (Figure 1B), adding corroborating support for these partitions. Further, comparative genomic analysis of sequenced isolates against the completed C. jeikeium K411 reference genome (Figure 2A) demonstrated variable degrees of divergence, and variable regions of divergence, among the clinical strains which seemingly partitioned isolates among the same groups identified by the genomic phylogeny (Figure 2B).
Figure 2. Circular plot of genome diversity in sequenced C. jeikeium clinical isolates.
A. The map, GC content, and GC skew (either positive [enrichment of G over C] or negative [enrichment of C over G]) of the fully sequenced C. jeikeium K411 reference genome is depicted in the innermost three rings. The white and colored regions of outer rings indicate sequences absent and present, respectively, in the draft genomes of clinical isolates and type strain ATCC 43734 relative to the C. jeikeium K411 reference. Intensity of coloration in outer rings indicates the degree of sequence identity relative to the reference genome. With the exception of the K411 reference strain, rings are grouped according to the genomospecies defined in Table 2. Strains are listed by genomospecies, from left to right, in the order that they occur when moving from the innermost to the outermost ring (key). B. Example of a divergent genomic region. Enlarged area is between 800 kbp and 1000 kbp in the K411 reference genome, as indicated. A pattern of divergence is observed that distinguishes group 1 (inner 9 rings) from other groups.
In order to formally circumscribe potential subdivisions among the isolates based on whole genome data, we performed an average nucleotide identity by BLAST (ANIb) analysis (Richter and Roselló-Móra, 2009) (Table 2). Pairwise ANIb values of less than 95% are generally accepted as the cutoff for defining separate species (Goris et al., 2007; Richter and Roselló-Móra, 2009), and by that criterion isolates in this study were sharply delineated into 4 separate species, which we have designated C. jeikeium genomospecies 1, 2, 3, and 4. The species most genomically divergent from existing reference strains (genomospecies 1) is predominant among the clinical isolates we have encountered, representing 9 of the 15 isolates included in this study. The other 3 genomospecies have fewer representatives: the group containing both existing C. jeikeium reference strains K411 and ATCC 43734 (genomospecies 2) bears two additional clinical isolates from our study, whereas the remaining two species (genomospecies 3 and 4) have only a single representative each.
Table 2.
Pairwise ANIb values based on whole genome data
| Genomospecies | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||||||||||||
| Isolate | Cj37130 | Cj30184 | Cj21382 | Cj14566 | Cj16348 | Cj38002 | Cj47453 | Cj19409 | Cj47446 |
C. jeikeium K411 |
C. jeikeium ATCC 43734 |
Cj47447 | Cj47445 | Cj30952 | Cj47444 | ||
| Genomospecies | 1 | Cj37130 | 100 | 97.78 | 97.69 | 97.55 | 97.43 | 97.76 | 97.14 | 97.64 | 97.14 | 85.87 | 85.63 | 85.87 | 86.01 | 86.04 | 86.02 |
| Cj30184 | 97.34 | 100 | 97.5 | 97.15 | 97.47 | 98.39 | 96.83 | 97.34 | 97.03 | 85.77 | 85.52 | 85.78 | 85.94 | 86.19 | 86.18 | ||
| Cj21382 | 97.6 | 97.71 | 100 | 97.43 | 97.54 | 97.51 | 96.96 | 97.59 | 97.26 | 85.75 | 85.86 | 85.77 | 85.84 | 86 | 85.92 | ||
| Cj14566 | 97.53 | 97.56 | 97.53 | 100 | 97.55 | 97.5 | 97.21 | 97.62 | 97.26 | 85.86 | 85.58 | 85.9 | 86.01 | 86.14 | 86.05 | ||
| Cj16348 | 97.36 | 97.65 | 97.49 | 97.46 | 100 | 97.44 | 97.03 | 97.55 | 97.24 | 85.83 | 85.77 | 86.14 | 86.1 | 85.99 | 86.09 | ||
| Cj38002 | 97.65 | 98.69 | 97.49 | 97.42 | 97.45 | 100 | 97.06 | 97.53 | 97.1 | 85.96 | 85.74 | 85.83 | 86.17 | 86.29 | 86.22 | ||
| Cj47453 | 96.96 | 96.95 | 96.98 | 96.94 | 96.96 | 96.84 | 100 | 97.14 | 96.59 | 86.11 | 85.85 | 86.12 | 86.26 | 86.25 | 86.08 | ||
| Cj19409 | 97.51 | 97.62 | 97.63 | 97.48 | 97.55 | 97.48 | 97.13 | 100 | 97.24 | 85.69 | 85.55 | 85.85 | 85.91 | 86.01 | 85.83 | ||
| Cj47446 | 97.13 | 97.01 | 97.06 | 97.21 | 97.07 | 97.07 | 96.52 | 97.06 | 100 | 86.31 | 86.07 | 86.5 | 86.6 | 86.24 | 86.45 | ||
| 2 | C. jeikeium K411 | 85.62 | 85.67 | 85.64 | 85.6 | 85.76 | 85.71 | 85.84 | 85.59 | 86.4 | 100 | 96.35 | 96.08 | 96.23 | 91.64 | 91.97 | |
| C. jeikeium ATCC 43734 | 85.28 | 85.42 | 85.66 | 85.28 | 85.56 | 85.48 | 85.48 | 85.37 | 85.73 | 96.47 | 100 | 96.45 | 96.71 | 91.55 | 91.43 | ||
| Cj47447 | 85.82 | 85.91 | 85.99 | 85.8 | 86.08 | 85.75 | 86.19 | 85.9 | 86.66 | 96.14 | 96.51 | 100 | 98.03 | 91.87 | 92.25 | ||
| Cj47445 | 85.83 | 85.86 | 85.87 | 85.88 | 85.94 | 86.06 | 86.09 | 85.86 | 86.75 | 96.28 | 96.61 | 97.92 | 100 | 91.84 | 92.53 | ||
| 3 | Cj30952 | 86.02 | 86.13 | 86.11 | 86.05 | 86.22 | 86.27 | 86.18 | 86.11 | 86.26 | 91.76 | 91.55 | 91.75 | 91.82 | 100 | 93.96 | |
| 4 | Cj47444 | 86.04 | 86.12 | 86.16 | 86.09 | 86.31 | 86.17 | 86.17 | 86 | 86.49 | 92.15 | 91.53 | 92.39 | 92.57 | 93.99 | 100 | |
Comparative gene content of C. jeikieum genomospecies
Differences among genomospecies were also manifested in their predicted gene content and core genome content (Table 3). Genomospecies 1 harbored significantly fewer predicted coding sequences (2-tailed t-test, p value = 0.009) and functionally-annotated coding sequences (2-tailed t-test, p value = 3.3×10−7) than isolates from genomospecies 2. The representative from genomospecies 3 had the fewest predicted coding sequences of any isolate, regardless, the number of functionally-annotated predicted coding sequences was within the range exhibited by members of genomospecies 2.
Table 3.
Comparative Gene Content of C. jeikeium genomospecies and core genome content
| Genomospecies | 1 | 2 | 3 | 4 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Cj37130 | Cj30184 | Cj21382 | Cj14566 | Cj16348 | Cj38002 | Cj47453 | Cj19409 | Cj47446 |
C. jeikeium K411 |
C. jeikeium ATCC 43734 |
Cj47447 | Cj47445 | Cj30952 | Cj47444 |
| Number Coding sequences | 2039 | 2136 | 1992 | 2063 | 2053 | 2036 | 2117 | 2080 | 2086 | 2211 | 2293 | 2148 | 2202 | 2001 | 2161 |
| Functionally-annotated Coding Sequences |
1382 | 1369 | 1374 | 1375 | 1365 | 1382 | 1378 | 1392 | 1374 | 1423 | 1435 | 1432 | 1434 | 1397 | 1452 |
| Functionally-annotated Coding sequences shared with K411 |
1362 | 1341 | 1355 | 1353 | 1347 | 1363 | 1358 | 1367 | 1351 | 1423 | 1401 | 1412 | 1408 | 1385 | 1408 |
| % K411 functionally-annotated coding sequences present |
95.71 | 94.24 | 95.22 | 95.08 | 94.66 | 95.78 | 95.43 | 96.06 | 94.94 | 100.00 | 98.45 | 99.23 | 98.95 | 97.33 | 98.95 |
| Estimated number of core genes for genomospecies |
1139 | 1384 | N/A* | N/A* | |||||||||||
Not applicable, only one representative of the genomospecies available.
The overlap in gene content between isolates and the fully-sequenced K411 reference strain mirrored the calculated degree of genomic divergence measured by ANIb: isolates from genomospecies 1 had the smallest percentage of predicted orthologous genes shared with K411 (Table 3), whereas members of genomospecies 2 had the highest. Although a core genome size could not be estimated for genomospecies 3 and 4, because these groups were represented by only a single isolate, the number of genes identified in isolate Cj47444 of genomospecies 4 which were not present in the sequenced reference strain (44 genes) was significantly higher than the number observed for other isolates (average of 21 genes, z-test p value = 1), suggesting a significant degree of additional genomic content for that genomospecies.
Phenotypic characterization of C. jeikieum genomospecies
We next investigated whether any of the genomospecies were distinguishable on the basis of phenotype. We performed a battery of standard biochemical tests used for the clinical classification of bacteria in patient samples, comprising 4 carbohydrate utilization tests, 5 glycosidase substrate tests, 5 aminopeptidase substrate tests, and 6 single-enzyme or phenotypic tests (Table 4). Isolate Cj30952, the sole representative of genomospecies 3, displayed a markedly different biochemical profile compared with other isolates, being uniquely characterized by an inability to utilize glucose and reduced aminopeptidase activity against leucine-β-naphthylamide. These characteristics separated this particular isolate from all other isolates in all other genomospecies. Metabolic profiles within and among the remaining genomospecies, however, were not distinct.
Table 4.
Biochemical testing results
| carbohydrate utilization |
glycosidase substrates | aminopeptidase substrates |
single enzyme or phenotypic tests |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomospecies | Isolate | glucose | sucrose | ribose | maltose | p-nitrophenyl-α-D-glycoside | p-nitrophenyl-β-D-glycoside | p-nitrophenyl-N-acetyl-βglycoside | p-nitrophenyl glycoside | o-nitrophenyl-β-d-galactopyranoside | proline-β-naphthylamide | tryptophan-β-naphthylamide | pyrrolidine-β-naphthylamide | leucyl-glycine-β-naphthylamide | leucine-β-naphthylamide | urease | nitrase reductase | catalase | yellow pigment | phosphatase | esterase |
| 1 | Cj37130 | + | −/+ | + | − | − | − | −/+ | − | − | − | + | − | + | + | − | − | + | − | + | + |
| Cj30184 | + | − | + | − | − | − | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| Cj21382 | + | − | + | − | − | + | − | − | − | −/+* | + | − | + | + | − | − | + | − | + | + | |
| Cj14566 | + | − | + | − | − | − | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| Cj16348 | + | − | + | − | − | − | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| Cj38002 | + | − | + | − | − | − | −/+ | − | − | −/+ | + | − | −/+ | + | − | − | + | − | + | + | |
| Cj47453 | + | − | + | − | − | − | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| Cj19409 | + | − | + | − | − | − | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| Cj47446 | + | − | + | − | − | −/+ | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| 2 | C. jeikeium K411 | + | − | − | − | − | −/+ | − | − | − | − | + | − | + | + | − | − | + | − | + | + |
|
C. jeikeium ATCC 43734 |
+ | − | −/+ | − | − | − | − | − | − | − | −/+ | − | + | + | − | − | + | − | + | + | |
| Cj47447 | + | − | + | − | − | − | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| Cj47445 | + | − | + | − | − | −/+ | − | − | − | − | + | − | + | + | − | − | + | − | + | + | |
| 3 | Cj30952 | − | − | − | − | − | − | − | − | − | − | −/+ | − | −/+ | −/+ | − | − | + | − | + | + |
| 4 | Cj47444 | + | − | + | − | − | −/+ | − | − | − | − | + | − | + | + | − | − | + | − | + | + |
weak positive
Similarly, we sought to determine whether the antibiotic susceptibilities of the genomospecies could be used to differentiate among them. All isolates tested were marked by resistance to penicillin and ceftriaxone, and were found to be either resistant or have intermediate resistance to clindamycin, and were sensitive to vancomycin (Table 5). Genomospecies 2 was uniquely distinguishable from other groups both in displaying resistance to gentamicin and also elevated resistance to tetracycline. Although not a uniquely defining characteristic, all members of genomospecies 2 also carried resistance to trimethoprim-sulfamethoxazole. Interestingly, although C. jeikeium strain K411 was initially identified as a multi-drug resistant strain from a hospitalized patient (Kerry-Williams and Noble, 1984), it appears that its resistance profile represents a general characteristic of genomospecies 2, rather than unique feature of the original K411 isolate. It was not possible to define other genomospecies based on their pattern of antibiotic susceptibilities: variable levels of resistance to ciprofloxacin and erythromycin were observed within genomospecies 1, and no consistent differences in resistance profiles distinguished among genomospecies 1, 3, and 4.
Table 5.
Antibiotic susceptibility profiles
| Gemnopecies | Isolate | Ceftriaxone | Ciprofloxacin | Clindamycin | Erythromycin | Gentamicin | Penicillin | Vancomycin | Tetracycline | Trimethoprim-sulfamethoxazole |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Cj37130 | 16# | 0.25 | >256 | 4 | 0.25 | 8 | 2 | 0.25 | 0.25/4.8 |
| Cj30184 | 16 | 0.25 | 2* | 0.5 | 0.125 | >32 | 1 | 0.5 | 1/19 | |
| Cj21382 | 16 | 0.25 | >256 | 8 | 0.125 | >32 | 2 | 0.5 | 0.5/9.5 | |
| Cj14566 | 32 | 0.25 | >256 | 32 | 0.125 | 8 | 2 | 0.5 | 1/19 | |
| Cj16348 | 16 | 8 | >256 | 8 | 0.125 | 16 | 1 | 0.5 | 0.5/9.5 | |
| Cj38002 | 32 | >32 | 4 | 0.25 | 0.25 | 8 | 2 | 0.5 | 0.125/2.4 | |
| Cj47453 | 8 | 0.25 | >256 | 4 | 2 | 8 | 2 | 0.5 | 1/19 | |
| Cj19409 | 16 | 0.125 | >256 | 16 | 0.25 | 4 | 1 | 0.5 | 1/19 | |
| Cj47446 | >32 | >32 | >256 | 64 | 1 | >32 | 2 | 2 | 8/152 | |
| 2 | C. jeikeium K411 | >32 | 0.5 | >256 | >256 | >1024 | >32 | 1 | 8* | >32/608 |
| C. jeikeium ATCC 43734 | >32 | 0.25 | 256 | 4 | >1024 | >32 | 2 | >256 | 16/304 | |
| Cj47447 | >32 | >32 | >256 | >256 | 512 | >32 | 1 | 64 | >32/608 | |
| Cj47445 | >32 | >32 | >256 | >32 | >1024 | >32 | 1 | 32 | >32/608 | |
| 3 | Cj30952 | >32 | >32 | >256 | >256 | 0.125 | >32 | 1 | 0.5 | 1/19 |
| 4 | Cj47444 | >32 | >32 | >256 | >256 | 0.125 | 32 | 1 | 0.5 | >32/608 |
All values reported in mg/L
Bold indicates full resistance,
indicates intermediate resistance
Discussion
The genus Corynebacterium, in general, is diverse and is notably difficult to speciate using phenotypic identification approaches (Roux et al., 2004), posing challenges for the task of distinguishing among isolates using standard biochemical and phenotypic classifiers. Further, available DNA-DNA hybridization data, although once considered a “gold standard” for circumscribing species, are somewhat difficult to interpret for C. jeikeium because the procedure utilized was noted to yield lower-than-average hybridization values (Grimont et al., 1980; Riegel et al., 1994). Advancements in genetic and whole genome sequencing technologies have now made it possible to infer intra-species population structure more accurately based on quantitative genomic information (Chan et al., 2012; Georgiades and Raoult, 2010; Richter and Roselló- Móra, 2009), enabling more comprehensive studies of Corynebacterium population structure. In this work we examined the genetic and genomic relationships among primary C. jeikeium clinical isolates identified through routine clinical workup in our laboratory, providing an unselected sampling of pathogenic strains collected over nearly a decade. Taken together, these data provide evidence that the 15 isolates considered here constitute 4 separate and distinct species.
Our findings are consistent with the earlier description of 4 “genomic groups” within the C. jeikeium taxon (Riegel et al., 1994), providing corroborating evidence for the conclusions presented in the present work. Nevertheless, our use of genomic sequencing data has enabled us to formally circumscribe these “groups” as true genomospecies based on quantitative metrics (Table 2), to precisely define the phylogenomic relationships among those genomospecies (Figure 1), and to explore the differences in genomic features that distinguishes them (Table 3). These investigations, coupled with phenotypic testing information and the generation of 13 new draft genome sequences derived from members of each genomospecies (Table 1), significantly expand knowledge of the genomic and phenotypic variation within the taxon. In light of these results, we advocate that C. jeikeium should be recognized as four separate species in order to accurately reflect the taxon’s true population structure.
Genomospecies 1, which comprises the majority of clinical isolates, has both a significantly smaller genome and significantly smaller gene repertoire than that of the other C. jekieum genomospecies (Table 3). Evidence suggests that the virulence potential of pathogenic bacteria is inversely proportional to overall genome size and gene content (Georgiades and Raoult, 2011), an observation which has been broadly demonstrated for phylogenetically dissimilar intracellular (Moran, 2002; Wixon, 2001) and extracellular (Georgiades and Raoult, 2011) disease-causing agents. Genome size itself has been shown to be a better predictor of bacterial virulence than the absolute number of classical “virulence factor” genes in comparing pathogenic bacteria compared to non-virulent or less-virulent counterparts (Georgiades and Raoult, 2011; Merhej et al., 2013), likely reflecting the process of gene loss during host adaptation (Merhej et al., 2009). Given this correlation, it is probable that genomospecies 1 is not overrepresented in our dataset due to chance sampling, but rather because it more frequently causes human disease than do the other genomospecies.
Of particular note, both of the extant C. jeikeium reference strains, K411 and ATCC 43734, belong to a single genomospecies (genomospecies 2), which is distinct from the clinically dominant genomospecies identified in our study (genomospecies 1). This finding has two major implications. First, it suggests that much of the genomic diversity present among members of the C. jeikeium taxon has not previously been represented through the two available reference strains. Secondly, it argues that studies of C. jeikeium pathogenesis may benefit from investigation of isolates classified to genomospecies 1, as existing C. jeikeium reference strains are not in a group responsible for the majority of infections encountered clinically.
Available data suggest that two of the four genomically-distinct species can be distinguished using routine clinical laboratory techniques: the sole representative of genomospecies 3 is uniquely characterized through negative glucose utilization (Table 4), while resistance to gentamicin can distinguish members of genomospecies 2 from others (Table 5). In the absence of additional testing, the only reliable primary means to classify representatives from genomospecies 1 and 4 is through the use of whole genome sequence analysis (Figure 1C). However, a biphasic approach should prove tractable to many clinical laboratories: first, phenotypic testing may be used to classify an unknown isolate to genomospecies 2 or 3 (Tables 4 and 5), followed as necessary by rpoB gene sequencing to differentiate among species 1 and 4 (Figure 1B).
Given that one genomospecies (genomospecies 1) has disproportionately high representation among the clinical isolates we have studied, and that two of the other genomospecies are poorly represented in this survey, we cannot know at this time whether we have fully sampled all species previously classified as C. jeikeium. The isolates examined here represent a comprehensive collection from our laboratory, but are nevertheless limited in absolute number and potentially geographical scope. It is possible that genomic characterization of supplementary isolates as they become available will delineate additional species, including those that may be less prevalent pathogens. Efforts should be taken to recognize and incorporate such groups into the Corynebacterium taxonomy, if they do indeed exist. Regardless, even now the proposed speciation of the C. jeikeium taxon carries with it important clinical diagnostic information about expected antibiotic resistance profiles of isolates classified to a particular group (Table 5). Future work may uncover additional clinically relevant distinctions among these species, such as preferred sites of infection or the severity of disease course caused by infection, shedding light on other potentially characterizing biological properties of these agents that are taxon-specific and not possible to assay by conventional laboratory testing.
Acknowledgment
This work was supported in part by the National Center For Advancing Translational Sciences of the National Institutes of Health [grant number UL1TR000423].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazanave C, Greenwood-Quaintance KE, Hanssen AD, Patel R. Corynebacterium prosthetic joint infection. J. Clin. Microbiol. 2012;50:1518–1523. doi: 10.1128/JCM.06439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JZ, Halachev MR, Loman NJ, Constantinidou C, Pallen MJ. Defining bacterial species in the genomic era: insights from the genus Acinetobacter. BMC Microbiol. 2012;12:302. doi: 10.1186/1471-2180-12-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarridge JE., 3rd Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clinical microbiology reviews. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke G, von Graevenitz A, Clarridge JE, 3rd, Bernard KA. Clinical microbiology of coryneform bacteria. Clinical microbiology reviews. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades K, Raoult D. Defining pathogenic bacterial species in the genomic era. Front. Microbiol. 2010;1:151. doi: 10.3389/fmicb.2010.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades K, Raoult D. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PloS one. 2011;6:e17962. doi: 10.1371/journal.pone.0017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Grimont PAD, Popoff MY, Grimont F, Coynault C, Lemelin M. Reproductibility and correlation study of three deoxyribonucleic acid hybridization procedures. Curr. Microbiol. 1980;4:325–330. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Ifantidou AM, Diamantidis MD, Tseliki G, Angelou AS, Christidou P, Papa A, Pentilas D. Corynebacterium jeikeium bacteremia in a hemodialyzed patient. International journal of infectious diseases. IJID. 2010;14(Suppl 3):e265–e268. doi: 10.1016/j.ijid.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Jackman PJH, Pitcher DG, Pelczynska S, Borman P. Classification of corynebacteria associated with endocarditis (group JK) as Corynebacterium jeikeium sp. nov. Syst. Appl. Microbiol. 1987;9:83–90. [Google Scholar]
- Kerry-Williams SM, Noble WC. Plasmid-associated bacteriocin production in a JK type coryneform bacterium. FEMS Microbiol. Lett. 1984;25:179–182. [Google Scholar]
- Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J. Clin. Microbiol. 2005;43:1934–1936. doi: 10.1128/JCM.43.4.1934-1936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup, 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Georgiades K, Raoult D. Postgenomic analysis of bacterial pathogens repertoire reveals genome reduction rather than virulence factors. Brief. Funct. Genomics. 2013;12:291–304. doi: 10.1093/bfgp/elt015. [DOI] [PubMed] [Google Scholar]
- Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol. direct. 2009;4:13. doi: 10.1186/1745-6150-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottumarthy S, Limaye AP, Prentice JL, Houze YB, Swanzy SR, Cookson BT. Nocardia veterana, a new emerging pathogen. J. Clin. Microbiol. 2003;41:1705–1709. doi: 10.1128/JCM.41.4.1705-1709.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Roselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegel P, de Briel D, Prévost G, Jehl F, Monteil H. Genomic diversity among Corynebacterium jeikeium strains and comparison with biochemical characteristics and antimicrobial susceptibilities. J. Clin. Microbiol. 1994;32:1860–1865. doi: 10.1128/jcm.32.8.1860-1865.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux V, Drancourt M, Stein A, Riegel P, Raoult D, La Scola B. Corynebacterium species isolated from bone and joint infections identified by 16S rRNA gene sequence analysis. J. Clin. Microbiol. 2004;42:2231–2233. doi: 10.1128/JCM.42.5.2231-2233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Kaiser O, Hain T, Goesmann A, Weisshaar B, Albersmeier A, Bekel T, Bischoff N, Brune I, Chakraborty T, Kalinowski J, Meyer F, Rupp O, Schneiker S, Viehoever P, Pühler A. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J. Bacteriol. 2005;187:4671–4682. doi: 10.1128/JB.187.13.4671-4682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tleyjeh IM, Qutub MO, Bakleh M, Sohail MR, Virk A. Corynebacterium jeikeium prosthetic joint infection: case report and literature review. Scand. J. Infect. Dis. 2005;37:151–153. [PubMed] [Google Scholar]
- Wixon J. Featured organism: reductive evolution in bacteria: Buchnera sp., Rickettsia prowazekii and Mycobacterium leprae. Comp. Funct. Genomics. 2001;2:44–48. doi: 10.1002/cfg.70. [DOI] [PMC free article] [PubMed] [Google Scholar]