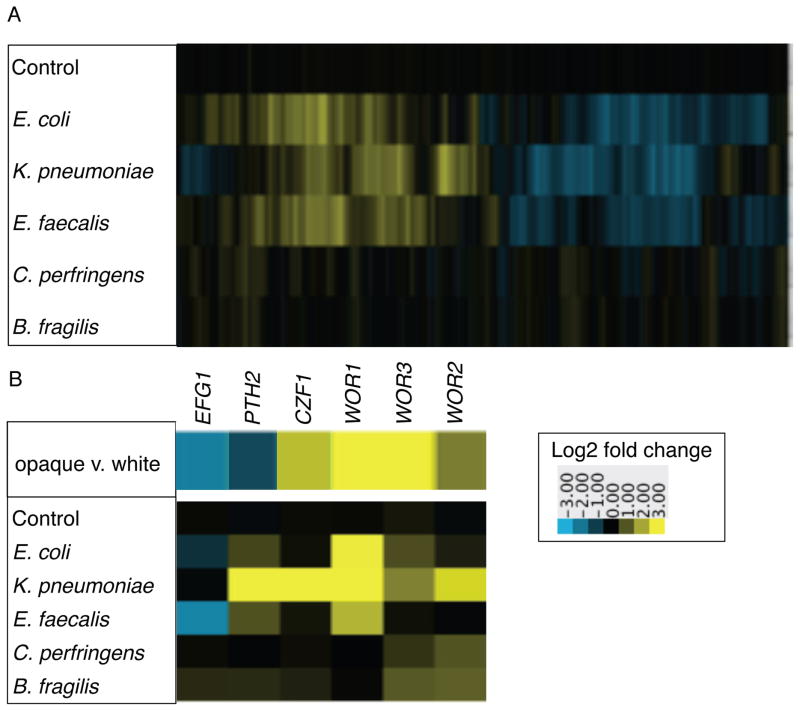
Figure 3. Co-culture with bacteria in biofilms induces differential gene expression in C. albicans.
A) Heat map of gene expression in C. albicans when co-cultured with the indicated species in biofilms, compared to C. albicans alone. Shown are the median values of at least two biological replicates. Control refers to C. albicans with media added to mimic the inoculum with bacteria, compared to C. albicans alone. 2863 genes differentially regulated at least twofold in at least one condition are displayed along the x-axis. Upregulated genes are yellow, downregulated genes are blue. B) Gene expression pattern of genes encoding transcription regulators that control the white-opaque switch circuit. The top panel shows expression levels measured in opaque vs. white cells from [19]. The bottom panel shows expression levels when C. albicans is co-cultured in biofilms with the indicated bacterial species, compared to C. albicans alone. See also Figure S3.