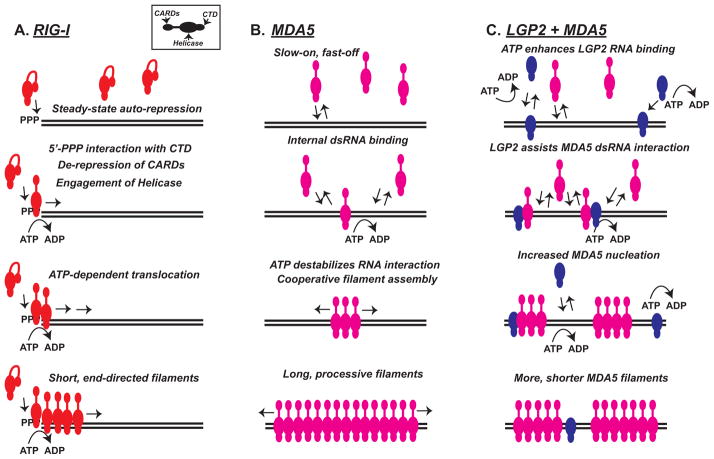
Figure 2. Comparison of RNA filament assembly by RLR proteins.
Panels illustrate features of RLR assembly onto RNA templates. Each RLR protein is illustrated as a colored module, and inset box demonstrates positions of CARD, Helicase and CTD regions. Black lines indicate dsRNA.
A) RIG-I (red) is auto-repressed at steady state by CARD-Hel2i interactions. Recognition of a 5′-triphosphorylated dsRNA terminus (5′-PPP) by the CTD results in CARD de-repression. The helicase domain mediates ATP-dependent translocation away from the RNA terminus with limited processivity, and leads to packaging of RIG-I into short, end-directed filaments.
B) MDA5 (magenta) has a low monomeric RNA binding affinity that is insensitive to terminal structures and characterized by a slow on-rate. ATP-mediated destabilization prevents MDA5 accumulation on dsRNA, but cooperative interactions between monomers can enhance processive filament assembly. Long MDA5 filaments are observed in vitro.
C) LGP2 (blue) has a high affinity for dsRNA and uses ATP hydrolysis to enable high-efficiency binding to diverse RNA species. LGP2 is able to facilitate and stabilize MDA5-dsRNA interactions leading to increased filament nucleation, producing a greater number of shorter MDA5 filaments.