Abstract
Manganese superoxide dismutase (MnSOD), a critical anti-oxidant enzyme, detoxifies the mitochondrial-derived reactive oxygen species, superoxide, elicited through normal respiration or the inflammatory response. Proinflammatory stimuli induce MnSOD gene expression through a eutherian-conserved, intronic enhancer element. We identified two prototypic enhancer binding proteins, TEAD1 and p65, that when co-expressed induce MnSOD expression comparable to pro-inflammatory stimuli. TEAD1 causes the nuclear sequestration of p65 leading to a novel TEAD1/p65 complex that associates with the intronic enhancer and is necessary for cytokine induction of MnSOD. Unlike typical NF-κB-responsive genes, the induction of MnSOD does not involve p50. Beyond MnSOD, the TEAD1/p65 complex regulates a subset of genes controlling the innate immune response that were previously viewed as solely NF-κB-dependent. We also identified an enhancer-derived RNA (eRNA) that is induced by either proinflammatory stimuli or the TEAD1/p65 complex, potentially linking the intronic enhancer to intra- and interchromosomal gene regulation through the inducible eRNA.
Introduction
Manganese superoxide dismutase (MnSOD) is the first line of defense against reactive oxygen species produced in normal cellular respiration and as a consequence of the inflammatory response [1]. MnSOD is a nuclear encoded, mitochondrial protein which catalyzes the rapid dismutation of superoxide radicals into molecular oxygen and hydrogen peroxide [2]. The pathophysiological relevance of MnSOD is highlighted by the phenotype of MnSOD−/− mice on two background strains: the first dies by P10 with dilated cardiomyopathy, liver/skeletal muscle steatosis, and metabolic acidosis [3]; while in the second strain, lethality occurs after P21 with derangement of myocardial mitochondria, degeneration of brainstem and basal ganglia neurons, severe anemia and progressive motor disturbances [4]. Of equal import, elevated MnSOD levels are cytoprotective by: suppressing the malignant phenotype in various cancer cell lines and tumor formation in xenograft and transgenic mouse models [5, 6]; protecting against beta-amyloid-induced neurodegeneration [7] and NMDA-mediated neuronal death [8]; inducing radioresistance [9, 10]; and blocking apoptosis from etoposide [11], IL-3 withdrawal [12], TNFα exposure [13], and TRAIL-mediated events [14].
We and others have demonstrated that proinflammatory mediators, e.g. TNF-α, IL-1β, and LPS, induce MnSOD expression through a proximal promoter [15–18] and intronic enhancer element [19, 20]. The proximal promoter of the rat MnSOD gene contains binding sites for 10 constitutively bound regulatory factors, as assessed by in vivo footprinting [15]. The transcription factor CEBPβ and the NF-κB complex are believed to interact with sites within the MnSOD enhancer [17, 20, 21]. In an attempt to identify and characterize functionally important cognate transcription factors associated with enhancer function, we employed a yeast one-hybrid assay. Our studies uncovered a previously unidentified pairing of transcription factors (TEAD1/p65) which form a novel complex, are bound to active chromatin and, most relevantly, cooperatively induce endogenous MnSOD gene expression. Classically, many genes regulated by the proinflammatory response were associated solely with NF-κB-dependent regulation. We showed in fact that the regulation of a subset of these genes is dependent on not only p65, but also its direct interaction with TEAD1. We also identified an enhancer-derived RNA (eRNA) [22] that is induced in response to either proinflammatory stimuli or by the TEAD1/p65 complex, potentially linking this eRNA to intra- and interchromosomal gene regulation.
EXPERIMENTAL PROCEDURES
Cell Culture and Transient Transfection
Rat pulmonary epithelial cells, L2, human alveolar adenocarcinoma cells, A549, and human lung fibroblast cells, HFL-1, were grown in complete Ham’s F12K medium (Life Technologies) supplemented with 10% fetal bovine serum, 10 μg/ml penicillin G, 0.1 mg/ml streptomycin, and 0.25 μg/ml amphotericin B at 37°C in humidified air with 5% CO2. For all transfections, 1.5 μg of DNA was added to 4.5 μl of FuGENE 6 (Roche) in serum-free medium to a final volume of 288 μl which was added to cells for 3 h, after which cells were rinsed with PBS and medium was refreshed. For transfection of hGH reporter constructs and overexpression studies, cells were batch-transfected with equimolar amounts of the indicated construct to control for transfection efficiency and seeded into separate plates 12 h after transfection and allowed to recover for 12 h, after which they were either left untreated or stimulated. L2 cells were utilized for all experiments unless otherwise noted.
Dimethyl Sulfide (DMS) in vivo Footprinting
In vivo footprinting was done as described [23] with modifications followed by ligation-mediated PCR and gel electrophoresis. Intact cells were untreated (control) or treated with DMS for 1–2 min, and DNA was purified by phenol/chloroform extraction and digested with BamH I followed by an additional phenol/chloroform extraction. DNA was then treated with piperidine and heated at 90°C to cleave modified guanine residues followed by ethanol precipitation and resuspension in TE. Samples were then subjected to ligation-mediated PCR (LMPCR). Briefly, following a first-strand synthesis step, a linker was ligated to the resulting cleaved fragment. This double stranded fragment was amplified with a primers to the linker and the BamH I cleavage site prior to visualization by genomic sequencing (Church and Gilbert, 1984).
Yeast One-hybrid Screening
Screening was done using tandem triplicate repeats of the MnSOD intronic enhancer fragment (5′-GGAAATTACCACATTCTGGAAATTTTAC-3′) in the Matchmaker yeast one-hybrid system (Clontech). Briefly, oligomeric fragments were cloned into pHISi or pLazZi vectors and transformed into the yeast YM4271 strain. Integration was confirmed through plating on auxotrophic media. Subsequent transformation of the library was confirmed similarly with drop-out media and increasing 3-AT (3-amino-1,2,4-triazole) concentrations and 108 cfu/ml were screened from a human brain library (Clontech).
Plasmid Construction and Mutagenesis
A 360 bp fragment spanning the rat MnSOD intronic enhancer from intron 2 was subcloned into the NdeI site of the human growth hormone (hGH) reporter plasmid and mutagenized by Quikchange mutagenesis. TEAD1, TEAD4 and p65 cDNA’s were subcloned from the pACT2 Clontech vector into the pcDNA3.1 mammalian expression vector. The p65 transactivation (TA) deletion and p50 constructs were kind gifts of Dr. Mary Waltner-Law (University of Florida). Myc-tagged constructs were created by subcloning the indicated cDNA into StrataClone Mammalian Expression Vectors (Stratagene).
RNA Isolation and Analysis
Total RNA was isolated as described by Chomczynski and Sacchi [24] with modifications [1] and analyzed by northern analysis and qRT-PCR. For northern analysis, RNA was size-fractionated on a 1% agarose-formaldehyde gel and electrotransferred to a charged nylon membrane (Zetabind, Cuno Laboratory Products) followed by UV cross linking. The membrane was hybridized with restriction digested randomly primed 32P-labeled probes using the Random Primers DNA labeling system (Gibco BRL). Membranes were hybridized and washed in 40 mM NaHPO4 pH 7.2, 1 mM EDTA, and 1% SDS. Membranes were analyzed by autoradiography. for qRT-PCR, total RNA was reverse-transcribed using the SuperScript™ first-strand synthesis kit (Invitrogen) with oligo(dT) or a gene-specific antisense MnSOD intron 2 primer for eRNA evaluation. 2 μl of the resulting cDNA was used for qPCR analysis using SYBRR Green Supermix with ROX™ (Bio-Rad Laboratories) in an ABI 7000 Sequence Detection System and analyzed by the ΔΔCT method normalized to cyclophilin A as described previously [10, 11]. Crossing threshold (CT) values from both cyclophilin A and respective target genes were used in the ΔΔCT method to calculate relative fold inductions [10, 11].
Chromatin Immunoprecipitation (ChIP)
ChIP was performed with the indicated antibodies and analyzed by gel electrophoresis or qPCR and quantified relative to input prior to immunoprecipitation. Briefly, cells were crosslinked with 1% formaldehyde for 10 min at 25°C and quenched with 125 mM glycine prior to chromatin isolation. The chromatin lysate was sonicated, centrifuged at 15,000×g for 5 min at 4°C, and immunoprecipitated overnight with the indicated antibodies; the no antibody controls being processed in the same manner lacking any antibodies. Resulting complexes were captured by incubation with either Protein A or G for polyclonal or monoclonal antibodies, respectively. Captured complexes were washed and eluted, and the DNA-protein cross-links were reversed at 65°C for 4 h. Purified DNA was then subjected to traditional PCR or qPCR with primers specific to the MnSOD intron 2 enhancer: (top strand: 5′-CAGGTCTGGGAAACGGGTTGAGTAATTG3′, bottom strand: 5′ GAGGAAAGTTGTCAGATGTCACCTTAGAGG3′), and the MnSOD promoter (top strand: 5′-CAAGGCGGCCCGAGAAGAGGCGGGG-3′; bottom strand: 5′-CTTGGACACA-GCTAGGCGCTGAC-3′. Traditional PCR fragments were fractionated on a 1.5% agarose gel, transferred to a nylon membrane, hybridized with a γ-ATP32 radiolabeled oligonucleotide specific to a region within the amplified region, and visualized by autoradioagraphy.
Immunoblot Analysis
Protein was isolated from treated cells in lysis buffer containing Complete™ Mini protease inhibitor (Roche) and the protein concentration was determined by a BCA (bicinchoninic acid) assay (Pierce). 20 μg of protein was separated on a 7.5% Tris/HCl Ready-Gel (Bio-Rad Laboratories) and electrotransferred on to a Trans-Blot® (Bio-Rad Laboratories) charged nitrocellulose membrane. Membranes were stained with Ponceau S (Sigma-Aldrich) to verify even loading. Membranes were blocked in 5% non-fat milk in TTBS, probed with the indicated antibodies, subjected to ECL (enhanced chemiluminescence) (GE Healthcare) and visualized by exposure to Hyperfilm™MP (GE Healthcare).
Immunoprecipitation
L2 cells were transfected with either TEAD1 and p65 or Flag-TEAD1 and myc-p65 at 65–70% confluency using FuGENE 6 (Roche). Protein was isolated 48 h post-transfection with RIPA lysis buffer and split into equal fractions One fraction received no antibody and the other was immunoprecipitated with either p65 or c-myc antibody for 1 h at 4°C. Complex capture was completed by the addition of 25 μl of Protein A/G beads and incubation for 1 h at 4°C. Samples were boiled in Laemelli buffer for 5 min and subjected to immunoblot analysis as described above.
Immunohistochemistry
L2 cells were grown on cover slips and treated with LPS (0.5 μg/ml) or IL-1β (2 ng/ml) for 0.5 h or transfected with the indicated construct. The cells were incubated for 24–48 h, washed twice with serum free media and fixed with 4% Paraformaldehyde for 10 min at 25°C. The cells were then washed once with serum free media, 3× with PBS, and additionally fixed with 100% methanol for 5 min at −20°C. The fixed cells were washed twice with PBS, blocked with 10% goat serum/PBS and incubated with primary antibodies overnight at 4°C. Secondary antibodies (Alexa fluorochrome conjugated anti-mouse FITC or anti-rabbit Texas Red) were incubated at 25°C for 1 h in the dark along with Hoescht dye at 1:1000 to stain nuclei. The cells were washed a final time with PBS, and the coverslips were mounted using the Prolong Antifade kit (Molecular Probes). The immunofluorescence analysis was performed on a Zeiss Axioplan 2 microscope using a 20× objective. For comparative pictures, gains were kept at the same level for primary pictures and secondary antibody only controls. For analysis of tagged proteins, fixation was done with 100% methanol for 5 min at −20°C; all remaining procedures were identical.
siRNA
L2 cells at ~40% confluency were transfected with a final concentration of 100 nM Dharmacon siRNAs for p65 using DharmaFECT 1 siRNA transfection reagent, and A549 cells were transfected with Qiagen siRNAs for TEAD1, TEAD2 and TEAD4 using Hiperfect transfection reagent. After 72 h, cells were collected and immunoblot and/or qRT-PCR analysis was used to assess protein knockdown, followed by qRT-PCR for each transcription factor or MnSOD.
Data Analysis
Densitometry of autoradiographs was performed using ImageJ software and qPCR was analyzed by the ΔΔCT method [10, 11]. Significance in three or more independent experiments was determined by a Student’s t-test as p≤0.05 (*) and p≤0.005 (**).
Results
In vivo footprinting analysis of the MnSOD intronic enhancer and identification of cognate transcription factors by yeast one-hybrid
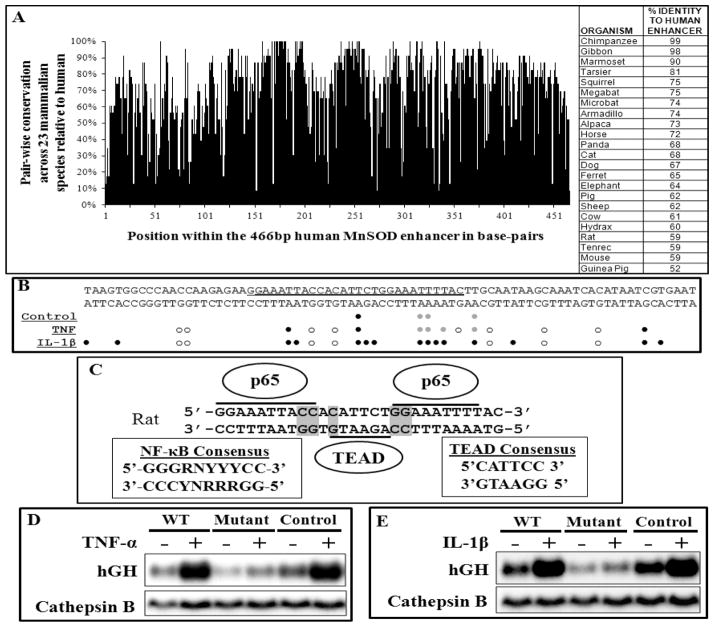
We and others identified a cytokine-dependent intronic enhancer region that controls MnSOD gene expression. Interestingly, this enhancer exhibits a high level of nucleotide identity over ~500 bp across mammalian species [19, 20, 25]. We provide a comparison of MnSOD intronic enhancer sequences from 23 eutherians, which by definition are placental mammals, compared to human (Fig. 1A). An in silico search using the 466 bp human MnSOD intronic fragment only found a high level of sequence identity amongst eutherians for which complete MnSOD gene sequences are available (Ensembl, WTSI/EBI) with no similar region identified in insect, fish, amphibian, reptile, bird, monotreme, or marsupial MnSOD genes.
Figure 1. Characterization of the MnSOD enhancer by in vivo footprinting, yeast one-hybrid and mutagenesis.
(A) Pair-wise conservation at each nucleotide across a 466 bp fragment of the human MnSOD intronic enhancer is based on a ClustalW alignment with 23 other eutherian enhancer regions. The adjacent table illustrates total percent identity of the enhancer for each eutherian species compared to human. (B) A summary of DMS in vivo footprinting results depicts control and stimulus-dependent (TNFα and IL-1β) enhancements (strong: ●, partial: ●) and protections (○) as compared to naked genomic DNA. (C) The underlined sequence in (B) was used for yeast one-hybrid analysis. The positive cDNAs from the one-hybrid screen were TEAD1, TEAD4 and p65. NF-κB and TEAD consensus binging sequences are provided. Two putative p65 sites and a TEAD site are marked with black bars on this region of the rat MnSOD intronic enhancer. Grey highlighted residues were mutated in functional studies. (D and E) Grey highlighted residues in (C) were mutated in a hGH reporter plasmid containing a rat MnSOD 360 bp enhancer fragment coupled to a 2.5 kb MnSOD promoter. Cells were transiently transfected with the wild type enhancer (WT), the mutated enhancer (Mutant) or an enhancer containing downstream mutations (Control), treated with TNFα (D) or IL-1β (E) for 8 h and analyzed by northern analysis. Cathepsin B serves as a loading control.
We previously demonstrated that MnSOD induction by proinflammatory mediators TNFα and IL-1β [1] requires this intronic enhancer element [19]. To identify the interaction of potential regulatory factors with this conserved intronic enhancer, we utilized dimethyl sulfate in vivo footprinting [15, 23] in a rat pulmonary epithelial cell line, L2, in the control and cytokine treated cells. We utilized rat L2 cells because all of our past studies on MnSOD, most importantly the previous characterization of the enhancer were performed in these cells. A summary of the in vivo footprinting data demonstrates protection (○), enhancement (●), or partial enhancement (●) of guanine and adenine residues indicating contacts with regulatory proteins (Fig. 1B). Specific contacts are observed that are unique to either TNFα or IL-1β treatment.
To identify cognate transcription factors that interact with the MnSOD enhancer, we utilized a yeast one-hybrid system to screen a human brain cDNA library to find relevant regulatory factors in a eukaryotic cellular setting. Figure 1C shows the rat sequence that we employed in the yeast one-hybrid screening. This screen identified three transcription factors, two of which are family members, transcriptional enhancer factor-1 (TEF1/TEAD1) and TEF3/TEAD4. TEAD1, the evolutionally conserved prototypic member of the family, was originally identified as the factor regulating the GT-IIC and Sph (I+II) enhansons of the simian virus 40 (SV40) enhancer [26]. The third cDNA identified from our one-hybrid screen encoded the gene for p65/RelA [27]. Historically, p65 functions as a homodimer or as a heterodimer with p105/50, classically defined as NF-κB. The canonical TEAD and p65 binding sequences are shown in Figure 1C. Residues within each of these putative binding sites were mutated in the context of a 360 bp rat MnSOD enhancer fragment in a human growth hormone (hGH) reporter plasmid also containing the 2.5 kb rat MnSOD promoter [19]. These constructs were transfected into L2 cells and evaluated in response to either TNFα or IL-1β by northern analysis, demonstrating the functional importance of these sequences (Figure 1D,E).
Analysis of TEAD1 and p65 enhancer binding and protein interaction
To evaluate the specificity of TEAD1 and p65 DNA binding, we generated TEAD1 and p65 glutathione-S-transferase (GST) fusion proteins which were expressed and isolated from E. coli using a glutathione affinity column. The purified proteins were used for EMSAs in the presence of nuclear extracts to assess enhancer specific DNA binding. Both proteins form shifted complexes with a fragment spanning the putative TEAD and p65 binding sites (Figures 1C and S1A). Specific binding of TEAD1 and p65 at these sites was confirmed by mutational analysis. Mutation of the central TEAD site (M-TEAD) eliminated all of the complexes (Figure S1A, middle). Analogously, the p65-GST protein forms two complexes, while a fragment where the 5′-most p65 site is mutated (M-p65) displays the loss of one shifted band with an intensification of the topmost band (Figure S1A, right), presumably through increased binding to the remaining 3′ p65 site (Figure 1C).
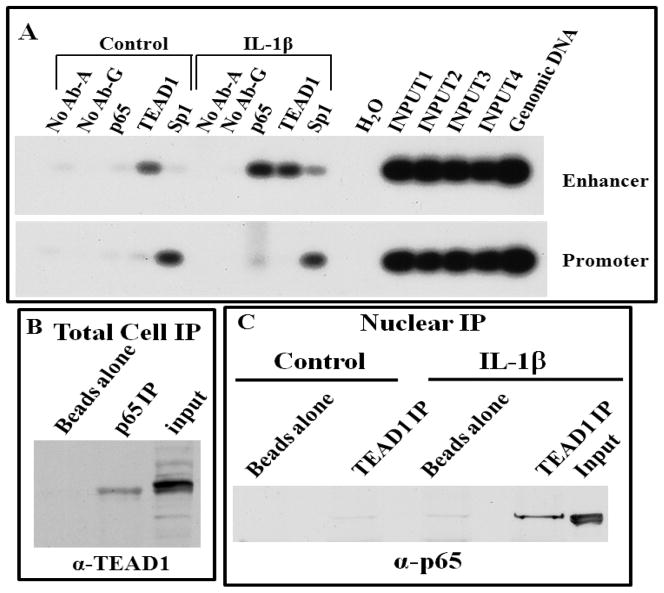
To further substantiate the results from the original yeast one-hybrid assay and verify the association of TEAD1 and p65 in mammalian cells with the intronic enhancer element, we utilized chromatin immunoprecipitation (ChIP) assays to identify protein-DNA interactions in native chromatin in L2 cells. A representative ChIP analysis for both the MnSOD intronic enhancer and promoter is shown in Figure 2A. As illustrated, TEAD1 is found at the enhancer in both the unstimulated and IL-1β-treated cells, while p65 is only found on the enhancer following IL-1β stimulation. We previously reported the constitutive interaction of the transcriptional activator Sp1 with three separate sites within the proximal MnSOD promoter by ChIP [16]. The results shown in Figure 2A confirm Sp1 interaction with the promoter region and demonstrate a reduced interaction of Sp1 with the enhancer region, similar to other studies [28]. As a negative control we also performed ChIP analysis utilizing primers flanking intron 3 and exon 4 (Figure S1B).
Figure 2. TEAD1 and p65 associate with the MnSOD enhancer and promoter by ChIP analysis and with each other by co-IP.
(A) Control and IL-1β (0.5 h) treated L2 cells were subjected to ChIP for p65, TEAD1 and Sp1. PCR of the enhancer or promoter regions was analyzed by Southern analysis. Protein A or G Sepharose beads served as the no antibody controls (No Ab-A/G), along with input DNA from each bead control (INPUT1–4) and non-sonicated genomic DNA. (B) Co-IP from total cell lysate from cells coexpressing TEAD1 and p65 using a p65 antibody followed by immunoblot analysis for TEAD1. (C) A reciprocal co-IP was performed on nuclear extracts from control and IL-1β-treated cells using a TEAD1 antibody followed by immunoblot analysis for p65. Total protein and beads alone serve as controls in (B, C).
To determine if TEAD1 and p65 interact directly with each other in cells, we employed co-immunoprecipitation (co-IP) studies. Figure 2B displays a total cellular p65-IP from L2 cells overexpressing both proteins followed by immunoblot for TEAD1, demonstrating an association between the two proteins in cells. To study stimulus-dependent nuclear interactions of these proteins, we used nuclear fractions from control and IL-1β-stimulated cells and performed a reciprocal co-IP using the TEAD1 antibody, confirming the stimulus-dependent formation of aTEAD1/p65 complex (Fig. 2C). These results are consistent with our ChIP results (Fig. 2A), demonstrating the presence of both proteins at the enhancer only after stimulation. To corroborate the specificity of the co-IP results, we overexpressed tagged proteins, FLAG-TEAD1, FLAG-TEAD4 and Myc-p65 in L2 cells followed by IP with a Myc antibody and immunoblotting with a FLAG antibody (Fig. S1C), confirming the presence of p65 in complexes with both TEAD1 and TEAD4.
TEAD1 sequesters p65 in the nucleus
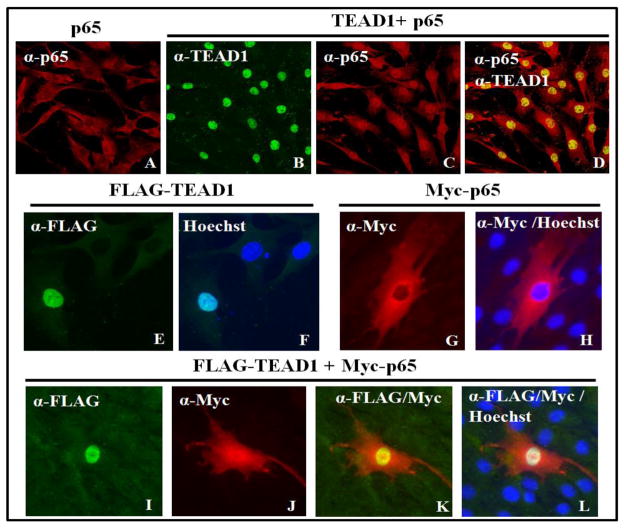
As part of the endogenous regulation of NF-κB activity, p65 homo- or heterodimers are sequestered in the cytosol through interaction with inhibitor of κB (IκB) subunits, with cellular stimulation releasing p65 and allowing translocation to the nucleus [29]. In control cells, endogenous p65 is found predominantly in the cytosol with TEAD1 exclusively nuclear as illustrated by immunoblot analysis of cellular fractions and immunohistochemistry (Fig. S2A–C). As documented, LPS or IL-1β treatment causes p65 translocation to the nucleus (Fig. S2A,D,E) [29]. Expression of p65 alone does not alter its cytosolic localization (Fig. 3A); however, coexpression of TEAD1 and p65 in L2 cells causes the efficient retention of p65 in the nucleus (Fig. 3B–D). To confirm these results, we overexpressed FLAG-TEAD1 and Myc-p65 alone or together (Fig. 3E–L). We also performed a similar experiment with the untagged proteins (Fig. S2F–L). These results demonstrate that TEAD1 is capable of sequestering p65 (Figs. 3, S2) in the nucleus through a complex containing these proteins (Figs. 2, S1). Our results are consistent with the ability of TEADs to mediate nuclear retention of the transcriptional activators YAP and TAZ, involved in the Hippo tumor suppressor pathway [30].
Figure 3. TEAD1 sequesters p65 in the nucleus based on immunohistochemistry.
(A) Immunohistochemistry of overexpressed p65 demonstrates cytosolic localization using a high DNA transfection. (B–D) Immunohistochemistry of cells overexpressing both p65 and TEAD1 show nuclear localization of TEAD1 (B) and p65 (C) and a merged image (D) with high DNA transfection. (E and F) Immunohistochemistry of overexpressed FLAG-tagged TEAD1 shows nuclear localization consistent with Hoechst staining with low DNA transfection. (G and H) Immunohistochemistry of overexpressed Myc-tagged p65 shows cytosolic localization as compared to Hoechst staining with low DNA transfection. (I–L) Immunohistochemistry of cells overexpressing both FLAG-tagged TEAD1 and Myc-tagged p65 shows nuclear localization of TEAD1 (I) and p65 (J) at a low DNA transfection with merged images for FLAG/Myc (K) and FLAG/Myc/Hoechst (L). All images were taken at 40× magnification with E–L digitally enlarged.
TEAD1 and p65 cooperatively induce endogenous MnSOD mRNA levels
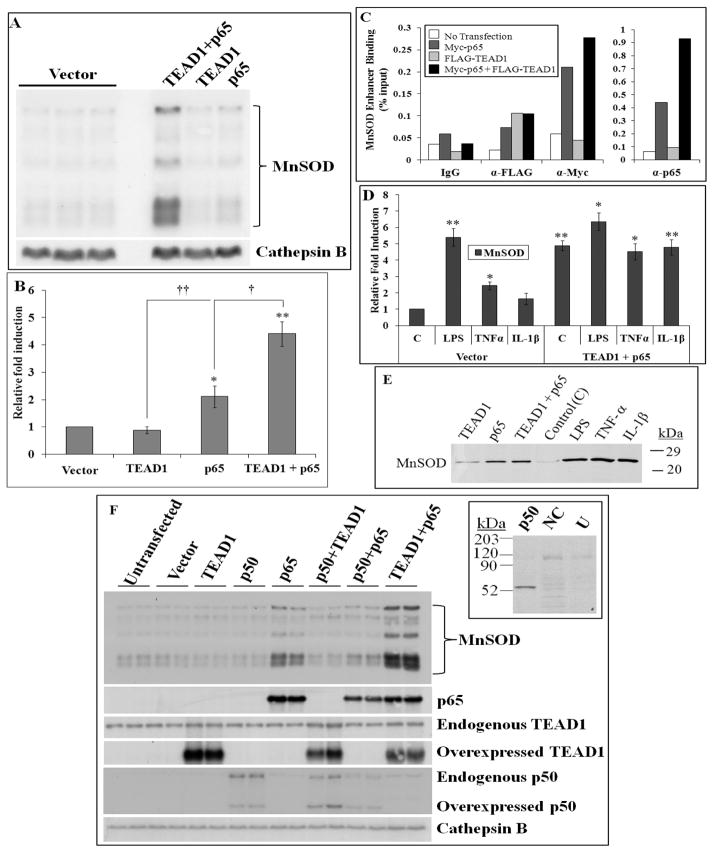
Having demonstrated that TEAD1 and p65 interact with each other and with the intronic MnSOD enhancer, we next tested the functional relevance of these interactions to endogenous MnSOD gene regulation. We previously showed that the rat MnSOD gene expresses five messages resulting from alternative polyadenylation [31]. A representative northern analysis shown in Figure 4A illustrates a striking induction of MnSOD mRNA levels when TEAD1 and p65 are coexpressed. p65 alone causes a small induction (~2×) while TEAD1/p65 results in a 4.4× induction of endogenous MnSOD mRNA levels (Fig. 4B). To further substantiate that these overexpressed proteins were directly affecting transcription through the enhancer, we overexpressed the tagged proteins, FLAG-TEAD1 and Myc-p65, and performed ChIP analysis with the respective Myc or FLAG antibodies followed by qPCR (Fig. 4C). As demonstrated, overexpression of p65 alone results in an increased association of the protein with the enhancer, while coexpression of TEAD1 and p65 potentiates an increased association, consistent with the northern analyses where coexpression of TEAD1 and p65 induces significantly higher MnSOD expression than p65 alone (Fig. 4A–C).
Figure 4. Overexpression of both TEAD1 and p65 induces MnSOD expression comparable to LPS and cytokine induction but independent of p50.
(A and B) Northern analysis (A) and corresponding densitometry of cells transfected with TEAD1, p65 or TEAD1+p65 compared to vector alone. Cathepsin B was used as a loading control in all northern analyses, n=6. (C) ChIP analysis at the MnSOD enhancer for FLAG, Myc or p65 was performed in cells overexpressing tagged proteins demonstrating increased association of p65 as a consequence of FLAG-TEAD1+Myc-p65 overexpression using qPCR. (D) Graphical summary of densitometry data from northern analyses of cells treated with LPS, TNFα, or IL-1β or in combination with transient transfection of TEAD1+p65. (E) Representative immunoblot analysis of MnSOD in L2 cells transfected with TEAD1, p65, or TEAD1+p65 for 48 h as compared to stimulation with LPS, TNFα, or IL-1β for 24 h. (F) Overexpression of p50 alone or in combination with TEAD1 or p65 does not induce MnSOD expression, as compared to p65 or TEAD1+p65. Inset demonstrates p50 overexpression, NC: negative control; U:untransfected. Significance in three or more independent experiments was determined by a Student’s t-test as p≤0.05 (*) and p≤0.005 (**) with data presented as mean ±SEM.
We next evaluated the effects of coexpression of TEAD1 and p65 within the context of stimulation with pro-inflammatory stimuli. As shown, combined overexpression of TEAD1 and p65 causes an induction of endogenous MnSOD mRNA comparable to that of LPS alone, which is not further enhanced by LPS, TNF-α, or IL-1β, implying that these mechanisms of induction contain overlapping components, most likely the transcription factors TEAD1 and p65 (Fig. 4D). Furthermore, the induction of MnSOD mRNA by TEAD1 and p65 is accompanied by increased MnSOD protein levels (Fig. 4E). As our yeast one-hybrid screen identified another TEAD family member, TEAD4, we performed comparable experiments with this factor. The TEAD1/4 family members alone or in combination had no effect on MnSOD expression (Fig. S3A,B); however, similar to the results seen for TEAD1, TEAD4 in combination with p65 also induces endogenous MnSOD mRNA to a level comparable to proinflammatory stimuli, implying redundancy of TEAD family members (Fig. S3C).
To further solidify the specificity of the TEAD/p65-mediated effects on gene expression, TEAD1 was coexpressed with C/EBPβ, another transcription factor we identified from a separate yeast one-hybrid screen that is also associated with cytokine-dependent induction of MnSOD [32]. TEAD1-C/EBPβ coexpression had no effect on MnSOD expression (Fig. S3D). To determine if our observations were specific to rat cells, we also tested the induction of MnSOD in response to TEAD1 and p65 in a human fetal lung fibroblast cell line, HFL-1, where coexpression elicited a similar induction of endogenous MnSOD (Fig. S3E).
The role of p50 in the regulation of MnSOD gene expression
To determine whether the classic component of NF-κB and dimerization partner for p65, p50 [29], has any involvement in MnSOD expression, p50 was expressed alone or in combination with TEAD1 or p65. p50 was unable to induce endogenous MnSOD in any of these conditions (Fig. 4F), despite protein overexpression significantly higher than endogenous levels (Fig. 4F, inset). As verification of functionality, overexpressed p50 causes an increase in its own endogenous p50 precursor mRNA (p105) (Fig. 4F), consistent with previous studies [33]. Therefore, p50 was undetectable in the control lanes because, similar to the immunoblot, the endogenous levels are low as compared to the overexpressed and induced endogenous p50 mRNA levels detected by northern analysis (Fig. 4F). To further evaluate any p50 contribution to stimulus-dependent MnSOD gene expression, we utilized a cell-permeable synthetic peptide inhibitor, SN50, that blocks nuclear translocation of the p50 subunit of NF-κB [34] (Fig. S3F). These data confirm the lack of contribution from p50 to the LPS, TNFα, or IL-1β inductions of MnSOD. Interestingly, p50 appears to inhibit the induction by p65 when they are coexpressed (Fig. 4F), and inhibition of p50 with SN50 accentuates the stimulus-dependent induction of specific rat MnSOD mRNA species (Fig. S4F).
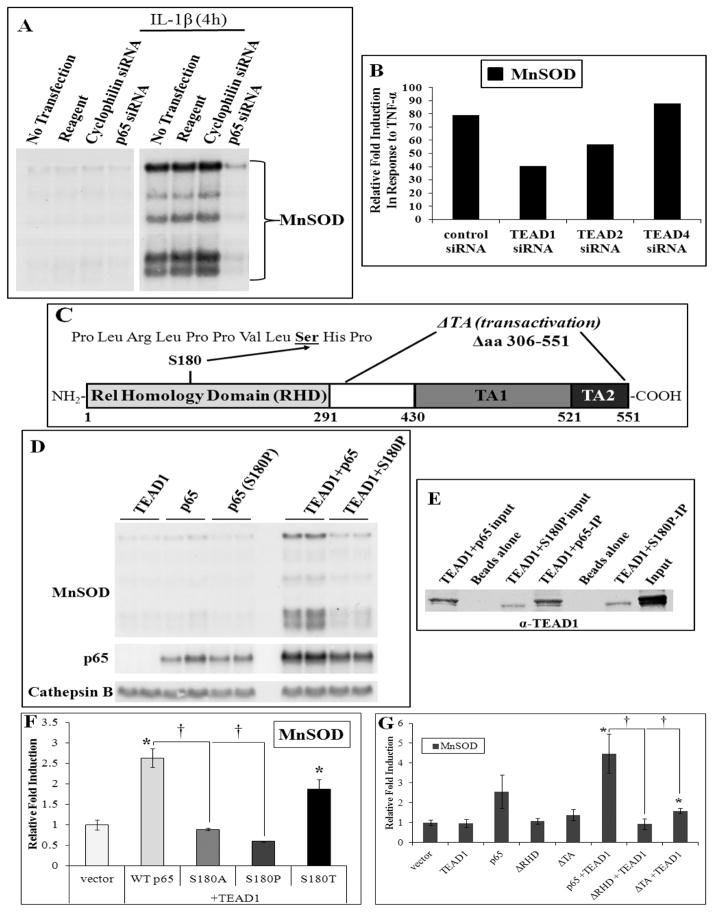
siRNA-mediated knockdown of p65 and TEAD1 inhibits cytokine induction of MnSOD
To evaluate the requirement of p65 or TEAD1 in cytokine-dependent stimulation of MnSOD, we first used siRNA knockdown of p65 in L2 cells stimulated with IL-1β. We confirmed knockdown of p65 protein levels by immunoblot (Fig. S4A). p65-specific siRNA effectively blocks induction of MnSOD mRNA levels in response to IL-1β signaling (Fig. 5A). TEAD1 and TEAD4 can interchangeably function in the induction of MnSOD (Figs. 4, S3) and redundant roles of TEAD1 and TEAD2 have previously been shown [35], therefore we analyzed the effect of knockdown of these three TEADs. Unfortunately, siRNAs were not available for all rat TEAD genes, therefore siRNA-mediated knockdown of human TEADs was performed in a human lung adenocarcinoma cell line, A549. Of note, A549 cells endogenously express equally high levels of TEAD1 and TEAD2, with lower TEAD4 levels (data not shown). Knockdown of each gene was verified by qRT-PCR, causing a 72%, 73% and 35% reduction of TEAD1, 2 and 4 mRNA, respectively (Fig, S4B). The induction of MnSOD mRNA in A549 cells by TNF-α was inhibited 48% and 28% by siRNA for TEAD1 and TEAD2, respectively, with no effect on induced MnSOD mRNA by a TEAD4 siRNA (Fig. 5B). The intermediate effectiveness of siRNA-mediated cytokine-dependent knockdown may be due to the inherent redundancy of the TEAD family [35] and the high level of affinity by each TEAD for the consensus sequence.
Figure 5. Cytokine induction of MnSOD requires TEAD1 and p65 along with serine 180 and the transactivation and Rel-homology domains of p65.
(A) siRNA knockdown of p65 inhibits IL-1β induction of MnSOD as compared to induction in the presence of transfection reagent or a control siRNA (cyclophilin B). (B) siRNA knockdown in A549 cells of TEAD1 or 2 partially inhibits TNFα-mediated induction of MnSOD with no effect by TEAD4 based on qRT-PCR analysis. (C) A diagram of the p65 protein coding region shows the location of serine 180 (S180) and the Rel homology (RHD) and two transactivation (TA1 and TA2) domains. (D) Representative northern analysis of p65 and p65 (S180P) proteins alone and coexpressed with TEAD1 demonstrating the importance of S180. (E) Immunoblot analysis of TEAD1 following p65 IP of cell extracts expressing WT p65 and the S180P mutant with TEAD1 demonstrating the ability of S180P to associate with TEAD1. (F) qRT-PCR analysis of MnSOD mRNA levels from total RNA isolated from cells coexpressing TEAD1 with WT p65, S180A, S180P and S180T, further demonstrating the importance S180 and reactivation based on threonine substitution. (G) Graphical summary of densitometry data from northern analyses of cells coexpressing WT p65, and deletions of the RHD (ΔRHD) or both TA domains (ΔTA) alone or in the presence of TEAD1 demonstrating the critical of importance of both regions. Significance in three or more independent experiments was determined by a Student’s t-test as p≤0.05 (* or †) with data presented as mean ±SEM.
The Rel homology domain and transactivation domains of p65 are required for cooperation with TEAD1
NF-κB transcriptional activity can be modulated through p65 phosphorylation in response to different stimuli [36, 37]. Prominent phosphorylation sites include S276, S529 and S536, which we have tested in the context of MnSOD regulation and found to not be involved (data not shown). We have however identified a serine residue in p65 at position 180 (S180, Fig. 5C) that, when mutated (S180P), abolishes the ability of p65 to induce MnSOD mRNA levels (Fig. 5D). The S180P protein was expressed in L2 cells (Fig. S4C) and, based on IP followed by immunoblot with anti-TEAD1 antibody, this mutant still binds to TEAD1, analogous to WT p65 (Fig. 5E). Mutation of S180 to threonine restores the MnSOD induction observed with WT p65 protein, supporting a hypothesis that this is indeed a phosphorylation site (Fig. 5F).
p65 contains two transactivation domains (TA1 and TA2) and a Rel homology domain (RHD) that contains the dimerization region and binding sites for IκB and DNA (Fig. 5C). To determine the relevance of theses domains to MnSOD induction, we expressed p65 constructs lacking either the RHD (ΔRHD) or the TA domains (ΔTA) (Fig. 5C) and verified expression by immunoblot and northern analysis (Fig. S4D,E). Northern analysis was used to verify expression of ΔRHD because the antibody epitope is deleted in this construct (Fig. S5E). The ΔRHD deletion, when coexpressed with TEAD1, is unable to elicit any induction of MnSOD, whereas the ΔTA construct with TEAD1 only causes a minor increase in MnSOD levels (Fig. 5G).
Stimulation of MnSOD transcription through the intronic enhancer occurs concurrently with induction of an enhancer-derived RNA (eRNA)
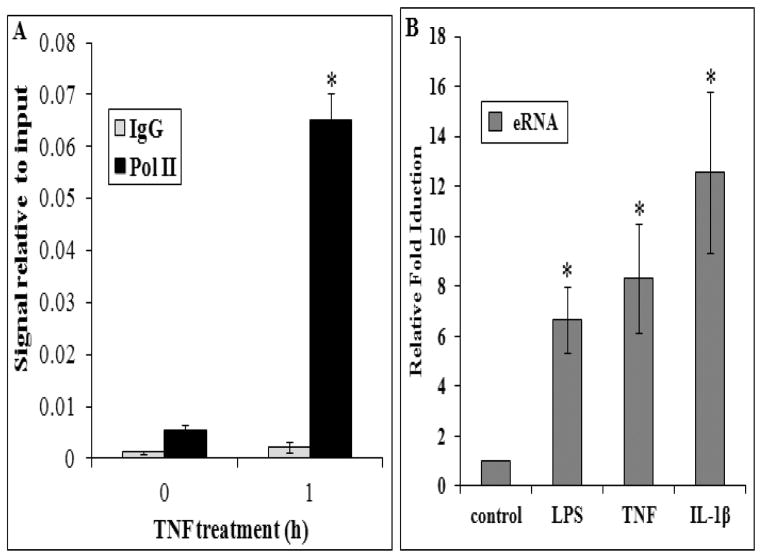
We previously demonstrated that both the human and rat MnSOD intronic enhancers drive stimulus-specific expression of a reporter construct, acting as its own promoter [19]. Since both the human and rat enhancer fragments were capable of initiating transcription from a site internal to the element in response to LPS, TNFα and IL-1β, we referred to this element as a prohancer [19]. Recently, Kim et al. [22] identified Pol II-dependent enhancer-associated transcripts which were bidirectional, relatively short, nonpolyadenylated, and in regions occupied by p300/CBP with high H3K4me1 but devoid of H3K4me3, calling these transcripts eRNAs. Similarly, genome-wide studies by De Santa et al. [38] showed that canonical enhancers represent 70% of extragenic Pol II localization. Enhancers are now considered to be the most common noncoding genomic elements undergoing transcription in higher eukaryotes. In 2013, several studies demonstrated a critical role of extragenic eRNAs in enhancer function [39–44]. Notably, Li et al. [43] and Hah et al. [39] showed that estrogen-activated ER-α causes genome-wide increases in eRNA transcription from enhancers controlling estrogen-regulated genes. These functional eRNAs act in long distance chromatin interactions by strengthening enhancer-promoter specific chromatin looping initiated by ER-α binding. Similarly, Melo et al. [41] demonstrated that p53-bound enhancers intrachromosomally regulate neighboring genes, requiring p53-dependent eRNAs to cause cell-cycle arrest. These elements are characterized by transcription of short eRNA messages that are necessary for transcriptional activation of the target gene(s). Due to the innate promoter activity of the MnSOD prohancer in a reporter construct, we hypothesized that this element may possess similar endogenous activity. We first tested whether RNA Pol II was inducibly bound to the MnSOD intronic enhancer. Within one hour of stimulation by TNFα, Pol II is recruited to this element (Fig. 6A). Using strand-specific reverse transcription reactions, we then analyzed the presence of an antisense eRNA transcript originating from the MnSOD enhancer. As shown in Figure 6B, LPS, TNFα and IL-1β all cause a robust induction of an antisense eRNA transcript within the enhancer. Due to the overlap of the sense strand eRNA with MnSOD intron and thus the homology to the unspliced heterogeneous nuclear RNA, we were unable to selectively study any sense eRNA transcription from the intronic enhancer. These results provide the first insights into inducible production of eRNA from an intronic or intragenic enhancer (Fig. 6B).
Figure 6. RNA Pol II inducibly associates with the MnSOD enhancer while LPS and cytokines stimulation causes the induction of an eRNA from the MnSOD enhancer.
(A) Control and TNFα (1 h) treated L2 cells were subjected to ChIP analysis for RNA Polymerase II (Pol II) at the MnSOD enhancer and samples analyzed by qPCR. (B) qRT-PCR of the enhancer-initiated eRNA from total RNA isolated from L2 cells treated with LPS, TNFα, or IL-1β for 8 h. Significance in three or more independent experiments was determined by a Student’s t-test as p≤0.05 (*) with data presented as mean ±SEM.
TEAD1 and p65 cooperate to induce genes previously associated with NF-κB
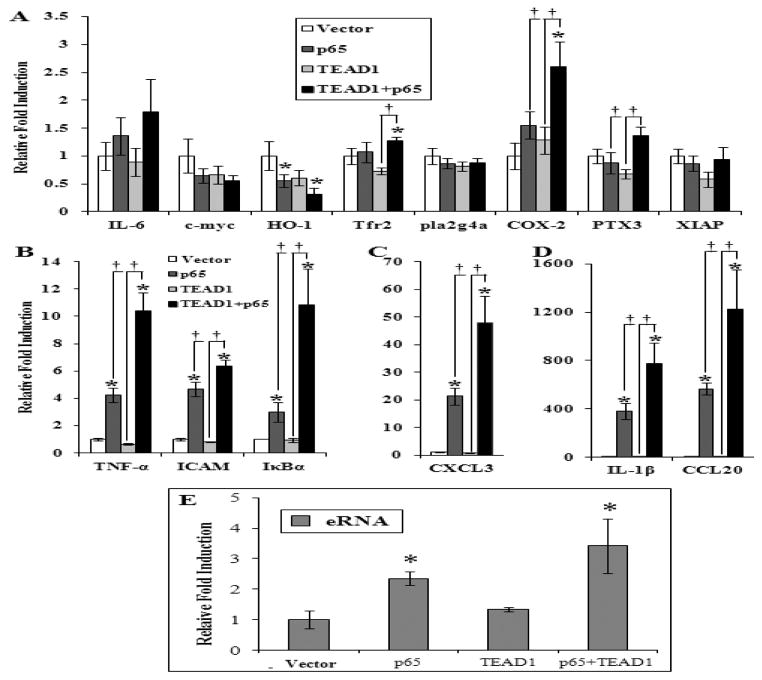
Having established TEAD1/p65, not p65/p50, as cooperative regulators of MnSOD, we sought to determine whether other classically NF-κB-associated genes were induced in response to these factors. Figure 7A illustrates qRT-PCR results for a number of previously reported NF-κB-regulated genes in response to TEAD1/p65 coexpression. Amongst these genes we observed only minor increases with Tfr2 and COX-2, whereas we observed a significant repression (69%) of the HO-1 gene in response to TEAD1/p65. While p65 alone could cause an induction of genes closely linked to the innate response, coexpression of TEAD1/p65 caused 10, 6, 11, 48, 774 and 1222× induction of the TNFα, ICAM, IκBα, CXCL3, IL-1β, and CCL20 genes, respectively (Fig. 7B–D). The selective effect of the unique pairing of TEAD1 and p65 implies that TEAD1 is a critical cofactor of p65 and the resulting complex regulates a subset of classical NF-κB-responsive genes.
Figure 7. TEAD1+p65 coexpression induces both classical NF-κB associated genes and the expression of eRNA from the MnSOD enhancer.
(A–D) qRT-PCR of the IL-6, c-Myc, heme oxygenase I (HO-1), tumor necrosis factor receptor 2 (TNFR2), group IV cytosolic phospholipase A2 (PLA2G4A), cyclooxygenase 2 (COX-2), pentraxin-related protein (PTX3), X-linked inhibitor of apoptosis protein (XIAP), TNF-α, Intercellular Adhesion Molecule 1 (ICAM-1), IκBα, chemokine C-X-C motif ligand 3 (CXCL3), IL-1β, and chemokine C-C motif ligand 20 (CCL20) genes from total RNA isolated from L2 cells expressing p65, TEAD1 or TEAD1+p65. (E) qRT-PCR of an enhancer initiated eRNA from total RNA isolated from L2 cells expressing p65, TEAD1 or TEAD1+p65. Significance in three or more independent experiments was determined by a Student’s t-test as p≤0.05 (* or †) with data presented as mean ± SEM.
Two potential mechanisms for the selective induction of these proinflammatory genes are gene-specific interactions of TEAD1/p65 or a TEAD1/p65-controlled master regulatory region which directly interacts with each of these genes. Individual eRNAs have been shown to interact with multiple genes to modulate their transcription [39–44]. If the MnSOD intronic enhancer is capable of functioning as one of these regions through its transcribed eRNA, then the eRNA must also be synthesized in response to TEAD1/p65 coexpression. Figure 7E demonstrates that this element produces an anti-sense eRNA in response to p65 which was more strongly activated in response to TEAD1/p65 coexpression. The induction of MnSOD (Fig. 4), other inflammatory genes (Fig. 7), and eRNA (Fig. 7) by p65 alone is consistent with the basal interaction of TEAD1 with the enhancer (Fig. 2) while elevated induction associated with coexpression results from increased co-occupancy. This effect implicates the MnSOD intronic enhancer as a potential mediator of broad proinflammatory regulation and thus innate immunity.
Discussion
MnSOD displays a high level of amino acid conservation from archaebacteria to humans [45], with a highly conserved intronic enhancer element critical for transcriptional regulation only present in eutherians. An analysis of insect, fish, amphibian, reptile, bird, monotreme, and marsupial MnSOD genes did not identify any homologous DNA element. As the only obvious features unique to eutherians, as compared to marsupials, are complex placentas and more fully developed offspring at birth, the basis for the recent evolution of this regulatory element in eutherians is not evident.
Our analysis of the enhancer includes a yeast one-hybrid screen identifying three proteins, TEAD1, TEAD4 and p65. These proteins were historically prototypic enhancer regulatory factors. TEADs were first isolated as part of a complex binding the SV40 enhancer region [26] whereas p65 was identified as part of a protein complex that bound the immunoglobulin enhancer regions [27]. Multiple studies have shown that TEAD1 can act as either a transcriptional inhibitor or activator [46–48] depending on the availability of suitable regulatory partners. TEADs exhibit functional redundancy [35] and lack transcriptional activation domains. TEADs facilitate DNA binding of multiple proteins including Max [49], PARP [50], SRF [51], TBP [52], and MEF2 [53]. The most notable TEAD partner is YAP [54], which, when complexed with TEAD1, controls cellular senescence [55], tissue growth [56], and tumorigenesis [57] through the SWH pathway [56, 58]. Our results demonstrate TEAD1-dependent sequestration of p65 in the nucleus where the novel TEAD1/p65 complex mediates cytokine-dependent regulation of MnSOD specifically through the intronic enhancer. Also supporting the involvement of TEAD1 in the regulation of MnSOD, mice deficient in either TEAD1 [35] or MnSOD [3, 4] display similar pathologies associated with cardiac myopathies.
p65 is a member of the evolutionarily and structurally conserved Rel homology domain family of proteins, which includes c-Rel, RelA (p65), RelB, p50/p105 and p52/p100 and has previously been implicated in MnSOD gene regulation [17, 21]. As part of the NF-κB complex, p65 is sequestered in the cytosol in most cells through an interaction with IκBs, a family of inhibitory molecules regulated through phosphorylation in response to inflammatory or stress-related stimuli. This phosphorylation releases the NF-κB complex and allows for nuclear import, DNA binding, and gene activation. The most heralded NF-κB complex consists of p50 and p65 [59]. Of the NF-κB subunits, p65 possesses the most potent activating potential through its C-terminal trans-activation domain [60]. However, our results have demonstrated that, unlike coexpression of TEAD1 and p65, the p50 subunit does not play a role in proinflammatory-mediated MnSOD gene regulation. As with TEAD1, new partners outside of the Rel family of proteins have been identified that may form relevant complexes with p65 [61, 62], and crystallization studies [63] have demonstrated an inherent flexibility in its ability to bind various DNA sequences. Here we show that a new partner for p65, TEAD1, functions to assist proinflammatory-mediated induction of MnSOD expression. Interestingly, both these proteins were initially argued to bind the SV40 enhancer region [27, 48] but at that time no correlation was made for their potential direct interaction and cooperation in enhancer function as we have demonstrated for MnSOD gene expression. We have also identified a novel serine residue (S180) in p65 that is required for MnSOD induction by cytokines and a S180T mutant supports this residue’s role as a potential phosphorylation site. Although the interaction and functional connection between TEAD1 and p65 has never been reported, they share a strikingly similar overlap in the control of physiological events including cellular senescence [55, 64], apoptotic resistance [65, 66], cell growth/differentiation [67, 68] and tumorigenesis [69, 70].
We therefore hypothesize that the TEAD1/p65 complex is not unique to MnSOD regulation, but also functions as an activator of other anti-apoptotic and anti-inflammatory genes. Our data demonstrate that TEAD1 and p65 regulate genes classically associated only with NF-κB, offering new insights into a critically important yet unrecognized regulatory mechanism. Remarkably, this complex causes significant induction of ICAM, TNFα, IL-1β, and the protein most closely linked to NF-κB regulation, IκBα. We also observe a substantial induction of the chemokines CXCL3 and CCL20. In addition, we also observe a TEAD1/p65-dependent repression (~69%) of the HO-1 gene. A specific linkage between p65 and HO-1 exists based on considerable evidence demonstrating suppression of NF-κB function through HO-1 anti-oxidant activity [71]. Moreover, a recent study has demonstrated that the Nrf2-ARE pathway is repressed through the interaction of p65 with Keap1 [72], thus potentially explaining the observed repression of HO-1 by TEAD1/p65. Collectively, these genes link the TEAD1/p65 complex to the control of the innate immune response. Many of these genes were associated with NF-κB based on sequence alone prior to technologies allowing for intracellular assessment of specific transcription factor binding and functional involvement. The advent of ChIP, ChIP-seq, and genome-wide transcriptional assays offer additional pursuits for studies involving genes coordinately-regulated by TEAD1 and p65.
The last three years of developments surrounding transcription from enhancer regions have uncovered a critical regulatory role for enhancer-derived non-coding RNAs. These enhancer elements encode small RNA transcripts referred to as eRNAs, critical to the transcriptional regulation of numerous intra- and interchromosomally localized target genes. Our discovery of transcription initiated from within the MnSOD intronic enhancer, first in a reporter construct [19] and endogenously in this study, establishes the MnSOD intronic enhancer as an eRNA-producing element. Most significantly, our data demonstrate that eRNA is transcribed in response to either proinflammatory stimuli (LPS, TNFα, or IL-1β) or the TEAD1/p65 complex. Given the distal regulation of many genes by eRNA and the TEAD1/p65-dependent regulation of this eRNA, we hypothesize that the TEAD1/p65-regulated genes identified here may be induced via this enhancer and associated eRNA.
To our knowledge, our data are the first evidence for the physical interaction and functional cooperation between TEAD1 and p65, prototypic enhancer binding proteins. This complex establishes an alternative mechanism to classic NF-κB signaling involved in the regulation of inflammatory and innate immune responses and substantiates a physiologically-relevant role for the TEAD1/p65 complex in the induction of gene expression by proinflammatory mediators. The identification of both the TEAD1/p65 complex and its associated eRNA defines a novel therapeutic target for the regulation of the inflammatory response and other common physiological events.
Supplementary Material
Acknowledgments
We would like to dedicate this manuscript to the memory of Joan Monnier who contributed greatly to these studies. H.S.N. was supported by NIH grants R37HL067456 and R01HL39593.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Visner GA, Dougall WC, Wilson JM, Burr IA, Nick HS. Regulation of manganese superoxide dismutase by lipopolysaccharide, interleukin-1, and tumor necrosis factor. Role in the acute inflammatory response. J Biol Chem. 1990;265:2856–2864. [PubMed] [Google Scholar]
- 2.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 3.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 4.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church SL, Grant JW, Ridnour LA, Oberley LW, Swanson PE, Meltzer PS, Trent JM. Increased manganese superoxide dismutase expression suppresses the malignant phenotype of human melanoma cells. Proc Natl Acad Sci U S A. 1993;90:3113–3117. doi: 10.1073/pnas.90.7.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong W, Oberley LW, Oberley TD, St Clair DK. Suppression of the malignant phenotype of human glioma cells by overexpression of manganese superoxide dismutase. Oncogene. 1997;14:481–490. doi: 10.1038/sj.onc.1200852. [DOI] [PubMed] [Google Scholar]
- 7.Keller JN, Kindy MS, Holtsberg FW, St Clair DK, Yen HC, Germeyer A, Steiner SM, Bruce-Keller AJ, Hutchins JB, Mattson MP. Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J Neurosci. 1998;18:687–697. doi: 10.1523/JNEUROSCI.18-02-00687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Zulueta M, Ensz LM, Mukhina G, Lebovitz RM, Zwacka RM, Engelhardt JF, Oberley LW, Dawson VL, Dawson TM. Manganese superoxide dismutase protects nNOS neurons from NMDA and nitric oxide-mediated neurotoxicity. J Neurosci. 1998;18:2040–2055. doi: 10.1523/JNEUROSCI.18-06-02040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eastgate J, Moreb J, Nick HS, Suzuki K, Taniguchi N, Zucali JR. A role for manganese superoxide dismutase in radioprotection of hematopoietic stem cells by interleukin-1. Blood. 1993;81:639–646. [PubMed] [Google Scholar]
- 10.Epperly MW, Chaillet JR, Kalash R, Shaffer B, Goff J, Franicola D, Zhang X, Dixon T, Houghton F, Wang H, Berhane H, Romero C, Kim JH, Greenberger JS. Conditional radioresistance of Tet-inducible manganese superoxide dismutase bone marrow stromal cell lines. Radiat Res. 2013;180:189–204. doi: 10.1667/RR3177.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantymaa P, Siitonen T, Guttorm T, Saily M, Kinnula V, Savolainen ER, Koistinen P. Induction of mitochondrial manganese superoxide dismutase confers resistance to apoptosis in acute myeloblastic leukaemia cells exposed to etoposide. Br J Haematol. 2000;108:574–581. doi: 10.1046/j.1365-2141.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- 12.Epperly MW, Bernarding M, Gretton J, Jefferson M, Nie S, Greenberger JS. Overexpression of the transgene for manganese superoxide dismutase (MnSOD) in 32D cl 3 cells prevents apoptosis induction by TNF-alpha, IL-3 withdrawal, and ionizing radiation. Exp Hematol. 2003;31:465–474. doi: 10.1016/s0301-472x(03)00041-9. [DOI] [PubMed] [Google Scholar]
- 13.Wong GH, Goeddel DV. Induction of manganous superoxide dismutase by tumor necrosis factor: possible protective mechanism. Science. 1988;242:941–944. doi: 10.1126/science.3263703. [DOI] [PubMed] [Google Scholar]
- 14.Mohr A, Buneker C, Gough RP, Zwacka RM. MnSOD protects colorectal cancer cells from TRAIL-induced apoptosis by inhibition of Smac/DIABLO release. Oncogene. 2008;27:763–774. doi: 10.1038/sj.onc.1210673. [DOI] [PubMed] [Google Scholar]
- 15.Kuo S, Chesrown SE, Mellott JK, Rogers RJ, Hsu JL, Nick HS. In vivo architecture of the manganese superoxide dismutase promoter. J Biol Chem. 1999;274:3345–3354. doi: 10.1074/jbc.274.6.3345. [DOI] [PubMed] [Google Scholar]
- 16.Kuo S, Chokas AL, Rogers RJ, Nick HS. PIN*POINT analysis on the endogenous MnSOD promoter: specific demonstration of Sp1 binding in vivo. Am J Physiol Cell Physiol. 2003;284:C528–534. doi: 10.1152/ajpcell.00356.2002. [DOI] [PubMed] [Google Scholar]
- 17.Maehara K, Hasegawa T, Xiao H, Takeuchi A, Abe R, Isobe K. Cooperative interaction of NF-kappaB and C/EBP binding sites is necessary for manganese superoxide dismutase gene transcription mediated by lipopolysaccharide and interferon-gamma. FEBS Lett. 1999;449:115–119. doi: 10.1016/s0014-5793(99)00408-1. [DOI] [PubMed] [Google Scholar]
- 18.Yeh CC, Wan XS, St Clair DK. Transcriptional regulation of the 5′ proximal promoter of the human manganese superoxide dismutase gene. DNA Cell Biol. 1998;17:921–930. doi: 10.1089/dna.1998.17.921. [DOI] [PubMed] [Google Scholar]
- 19.Rogers RJ, Chesrown SE, Kuo S, Monnier JM, Nick HS. Cytokine-inducible enhancer with promoter activity in both the rat and human manganese-superoxide dismutase genes. Biochem J. 2000;347(Pt 1):233–242. [PMC free article] [PubMed] [Google Scholar]
- 20.Jones PL, Ping D, Boss JM. Tumor necrosis factor alpha and interleukin-1beta regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-beta and NF-kappaB. Mol Cell Biol. 1997;17:6970–6981. doi: 10.1128/mcb.17.12.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maehara K, Hasegawa T, Isobe KI. A NF-kappaB p65 subunit is indispensable for activating manganese superoxide: dismutase gene transcription mediated by tumor necrosis factor-alpha. J Cell Biochem. 2000;77:474–486. doi: 10.1002/(sici)1097-4644(20000601)77:3<474::aid-jcb12>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 22.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, St Clair DK. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 1999;18:709–722. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- 26.Davidson I, Xiao JH, Rosales R, Staub A, Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988;54:931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- 27.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 28.Guo Z, Boekhoudt GH, Boss JM. Role of the intronic enhancer in tumor necrosis factor-mediated induction of manganous superoxide dismutase. J Biol Chem. 2003;278:23570–23578. doi: 10.1074/jbc.M303431200. [DOI] [PubMed] [Google Scholar]
- 29.Napetschnig J, Wu H. Molecular basis of NF-kappaB signaling. Annu Rev Biophys. 2013;42:443–468. doi: 10.1146/annurev-biophys-083012-130338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurt J, Hsu JL, Dougall WC, Visner GA, Burr IM, Nick HS. Multiple mRNA species generated by alternate polyadenylation from the rat manganese superoxide dismutase gene. Nucleic Acids Res. 1992;20:2985–2990. doi: 10.1093/nar/20.12.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu X, Aiken KJ, Chokas AL, Beachy DE, Nick HS. Distinct functions of CCAAT enhancer-binding protein isoforms in the regulation of manganese superoxide dismutase during interleukin-1beta stimulation. J Biol Chem. 2008;283:25774–25785. doi: 10.1074/jbc.M801178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ten RM, Paya CV, Israel N, Le Bail O, Mattei MG, Virelizier JL, Kourilsky P, Israel A. The characterization of the promoter of the gene encoding the p50 subunit of NF-kappa B indicates that it participates in its own regulation. Embo J. 1992;11:195–203. doi: 10.1002/j.1460-2075.1992.tb05042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin YZ, Yao SY, Veach RA, Torgerson TR, Hawiger J. Inhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J Biol Chem. 1995;270:14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- 35.Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeulen L, De Wilde G, Notebaert S, Vanden Berghe W, Haegeman G. Regulation of the transcriptional activity of the nuclear factor-kappaB p65 subunit. Biochem Pharmacol. 2002;64:963–970. doi: 10.1016/s0006-2952(02)01161-9. [DOI] [PubMed] [Google Scholar]
- 37.Hochrainer K, Racchumi G, Anrather J. Site-specific phosphorylation of the p65 protein subunit mediates selective gene expression by differential NF-kappaB and RNA polymerase II promoter recruitment. J Biol Chem. 2013;288:285–293. doi: 10.1074/jbc.M112.385625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He H, Li W, Wu D, Nagy R, Liyanarachchi S, Akagi K, Jendrzejewski J, Jiao H, Hoag K, Wen B, Srinivas M, Waidyaratne G, Wang R, Wojcicka A, Lattimer IR, Stachlewska E, Czetwertynska M, Dlugosinska J, Gierlikowski W, Ploski R, Krawczyk M, Jazdzewski K, Kere J, Symer DE, Jin V, Wang Q, de la Chapelle A. Ultra-rare mutation in long-range enhancer predisposes to thyroid carcinoma with high penetrance. PLoS One. 2013;8:e61920. doi: 10.1371/journal.pone.0061920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 42.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.May BP, Dennis PP. Evolution and regulation of the gene encoding superoxide dismutase from the archaebacterium Halobacterium cutirubrum. J Biol Chem. 1989;264:12253–12258. [PubMed] [Google Scholar]
- 46.Chaudhary S, Brou C, Valentin ME, Burton N, Tora L, Chambon P, Davidson I. A cell-specific factor represses stimulation of transcription in vitro by transcriptional enhancer factor 1. Mol Cell Biol. 1994;14:5290–5299. doi: 10.1128/mcb.14.8.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang SW, Trujillo MA, Eberhardt NL. Human chorionic somatomammotropin enhancer function is mediated by cooperative binding of TEF-1 and CSEF-1 to multiple, low-affinity binding sites. Mol Endocrinol. 1997;11:1223–1232. doi: 10.1210/mend.11.9.9984. [DOI] [PubMed] [Google Scholar]
- 48.Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 49.Gupta MP, Amin CS, Gupta M, Hay N, Zak R. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac alpha-myosin heavy-chain gene expression. Mol Cell Biol. 1997;17:3924–3936. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butler AJ, Ordahl CP. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol. 1999;19:296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta M, Kogut P, Davis FJ, Belaguli NS, Schwartz RJ, Gupta MP. Physical interaction between the MADS box of serum response factor and the TEA/ATTS DNA-binding domain of transcription enhancer factor-1. J Biol Chem. 2001;276:10413–10422. doi: 10.1074/jbc.M008625200. [DOI] [PubMed] [Google Scholar]
- 52.Jiang SW, Eberhardt NL. TEF-1 transrepression in BeWo cells is mediated through interactions with the TATA-binding protein, TBP. J Biol Chem. 1996;271:9510–9518. doi: 10.1074/jbc.271.16.9510. [DOI] [PubMed] [Google Scholar]
- 53.Maeda T, Chapman DL, Stewart AF. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J Biol Chem. 2002;277:48889–48898. doi: 10.1074/jbc.M206858200. [DOI] [PubMed] [Google Scholar]
- 54.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Q, Chen J, Feng H, Peng S, Adams U, Bai Y, Huang L, Li J, Huang J, Yuan Z, Meng S. YAP/TEAD-mediated transcription controls cellular senescence. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-3793. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Grusche FA, Harvey KF. Control of tissue growth and cell transformation by the Salvador/Warts/Hippo pathway. PLoS One. 2012;7:e31994. doi: 10.1371/journal.pone.0031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hilman D, Gat U. The evolutionary history of YAP and the hippo/YAP pathway. Mol Biol Evol. 2011;28:2403–2417. doi: 10.1093/molbev/msr065. [DOI] [PubMed] [Google Scholar]
- 59.Dyson HJ, Komives EA. Role of disorder in IkappaB-NFkappaB interaction. IUBMB Life. 2012;64:499–505. doi: 10.1002/iub.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitz ML, Baeuerle PA. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. Embo J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stein B, Baldwin AS, Jr, Ballard DW, Greene WC, Angel P, Herrlich P. Cross-coupling of the NF-kappa B p65 and Fos/Jun transcription factors produces potentiated biological function. Embo J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein B, Cogswell PC, Baldwin AS., Jr Functional and physical associations between NF-kappa B and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen YQ, Sengchanthalangsy LL, Hackett A, Ghosh G. NF-kappaB p65 (RelA) homodimer uses distinct mechanisms to recognize DNA targets. Structure. 2000;8:419–428. doi: 10.1016/s0969-2126(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Jacob NK, Ladner KJ, Beg A, Perko JD, Tanner SM, Liyanarachchi S, Fishel R, Guttridge DC. RelA/p65 functions to maintain cellular senescence by regulating genomic stability and DNA repair. EMBO Rep. 2009;10:1272–1278. doi: 10.1038/embor.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Landin Malt A, Cagliero J, Legent K, Silber J, Zider A, Flagiello D. Alteration of TEAD1 expression levels confers apoptotic resistance through the transcriptional up-regulation of Livin. PLoS One. 2012;7:e45498. doi: 10.1371/journal.pone.0045498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 67.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joyce D, Albanese C, Steer J, Fu M, Bouzahzah B, Pestell RG. NF-kappaB and cell-cycle regulation: the cyclin connection. Cytokine Growth Factor Rev. 2001;12:73–90. doi: 10.1016/s1359-6101(00)00018-6. [DOI] [PubMed] [Google Scholar]
- 69.Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- 70.Bharti AC, Aggarwal BB. Nuclear factor-kappa B and cancer: its role in prevention and therapy. Biochem Pharmacol. 2002;64:883–888. doi: 10.1016/s0006-2952(02)01154-1. [DOI] [PubMed] [Google Scholar]
- 71.Nakajima S, Kitamura M. Bidirectional regulation of NF-kappaB by reactive oxygen species: a role of unfolded protein response. Free Radic Biol Med. 2013;65:162–174. doi: 10.1016/j.freeradbiomed.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 72.Yu M, Li H, Liu Q, Liu F, Tang L, Li C, Yuan Y, Zhan Y, Xu W, Li W, Chen H, Ge C, Wang J, Yang X. Nuclear factor p65 interacts with Keap1 to repress the Nrf2-ARE pathway. Cell Signal. 2011;23:883–892. doi: 10.1016/j.cellsig.2011.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.