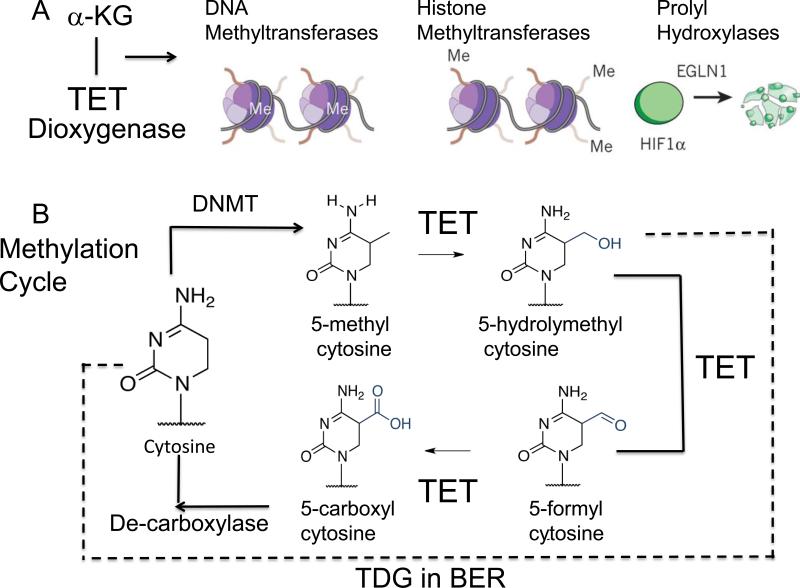
Figure 4. The Methylation cycle and its metabolic regulation.
(A) α-ketoglutarate (α KG) is a critical co-factor for the Ten-eleven translocation dioxygenase (TET) which oxidizes 5-methylcytosine. TET regulates both DNA and histone methylation, and EGLN1 gene product, hypoxia-inducible factor prolyl hydroxylase 2 (HIF-PH2), or prolyl hydroxylase domain-containing protein 2 (PHD2), is an enzyme encoded by the EGLN1 gene. It is also known as Egl nine homolog 1.The EGLN1 gene is responsible for ubiquitin-proteasome degradation pathway by hydroxylation of proline-564 and proline-402 by PHD2. (B) The methylation cycle regulated by TET. The action of DNA demethytransferases (DNMTs) converts cytosine (C) for 5’ methyl cytosine. The TET enzymes oxidize 5’ methyC to 5’-hydroxylmethyC and 5’formylC. The 5’-hydroxylmethyC is removed by the BER enzyme, TDG, to restore C, which is subsequently methylated. 5’formylC is caboxylated by an unknown enzyme to restore C, which is subsequently methylated. Success of this cycle maintains a balance of methylation in the genome. The dotted line indicates the removal of 5-hydrolymethylcytosine and restoration to cytosine to re-set the cycle.