Figure 1.
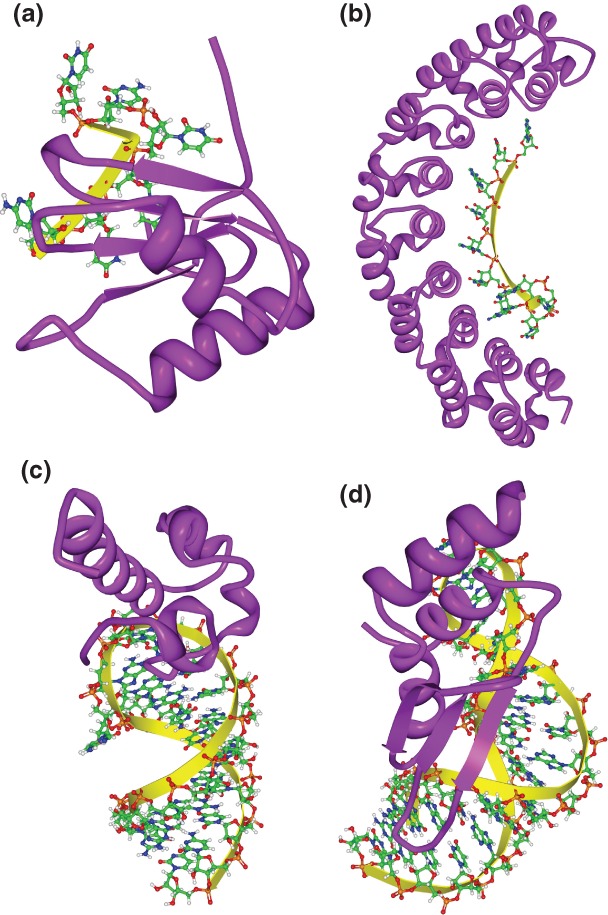
Three-dimensional structures of RNA-binding domain (RBD)–RNA complexes. (a) Solution structure of polypyrimidine tract binding (PTB) protein RBD1 in complex with CUCUCU RNA [Protein Data Bank (PDB): 2AD9]. PTB RBD1 binds a YCU site (Y indicating pyrimidine) through β4, β1, and β2, respectively. (b) Co-crystal structure of the PUM-homology domain (PUM-HD) in human Pum1 complexed with a 10-nucleotide single-stranded RNA, 5′-AUUGUACAUA where the last eight nucleotides (UGUACAUA) are individually recognized by three conserved amino acids in Puf repeats 8 to 1, respectively14 (PDB: 1M8Y). (c) Solution structure of the Vts1p sterile-α motif (specific affinity matrix, SAM) domain in complex with a 5′-CUGGC-3′ pentaloop as part of a 19nt hairpin (PDB: 2ESE). The specific interaction between the Vts1p SAM domain and the target RNA is stabilized by both the direct interaction to the third guanosine base in the RNA pentaloop and the contacts to the unique backbone structure.16–18 (d) Solution structure of dsRBD of yeast Rnt1p in complex with the 5′ terminal AGNN tetraloop of snR47 precursor RNA (PDB: 1T4l). Neither A nor G are recognized by specific hydrogen bonds; instead, the N-terminal helix of the Rnt1p dsRBD interacts with the backbone and the two nonconserved tetraloop bases, by snugly fitting into the minor groove side of the RNA tetraloop and extending into the minor groove at the top of the stem.19
