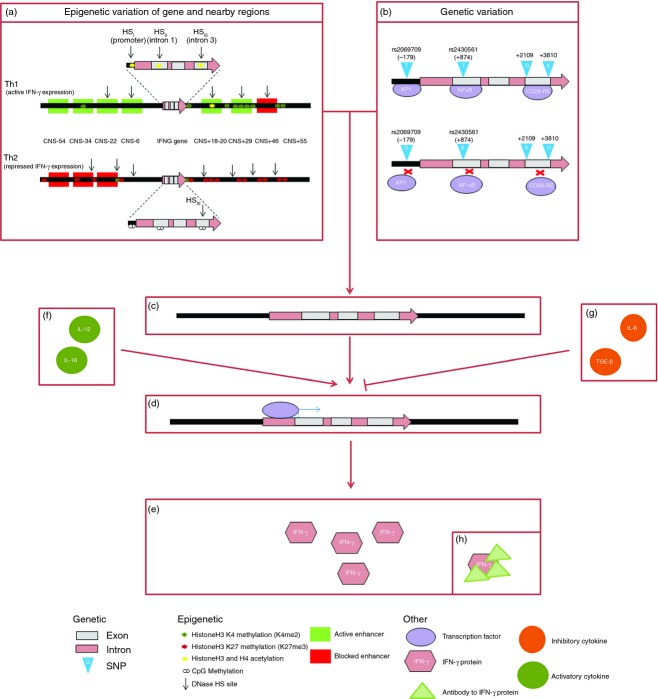
Figure 2.
Differential mechanisms to impairment of interferon-γ responses. Epigenetic (a) and genetic (b) variations affect the chromatin structure and result in a specific pattern of variation at the DNA level of the interferon-γ gene (IFNG) and surrounding regions (c). Epigenetic variations include histone acetylation and H3K4 methylation, which activate gene expression, and DNA CpG methylation and histone H3K27 methylation, which repress gene expression. Genetic variation includes single nucleotide polymorphisms (SNPs) that affect the presence or functioning of a transcription factor binding site. Rs2430561 (+874T/A) is located in a nuclear factor-κB (NF-κB) binding site and NF-κB binds preferentially to the T allele. Rs2069709 (−179G/T) is located in the promoter. The T may create a putative activator protein 1 (AP-1) binding element or oestrogen-like response element. SNPs in intron 3 (+2109A/G and +3810G/A) may also affect transcription, by altering the binding of protein complexes such as CD28-RE. Binding of transcription factors (d) is affected by this genetic and epigenetic variation as well as by the presence or absence of the transcription factors themselves, and binding of different transcription factors activates or represses expression of the IFNG gene, to affect production of IFN-γ protein (e). Cytokines such as interleukin-12 (IL-12) and IL-18 can promote IFNG expression (f), while those such as IL-6 and transforming growth factor-β (TGF-β) can prevent expression (g), so altering the production of IFN-γ protein (e). This IFN-γ protein can be prevented from functioning by the presence of IFN-γ antibodies (h).