Figure 8. MEF2A directly regulates basal Mfn2 expression in neurons.
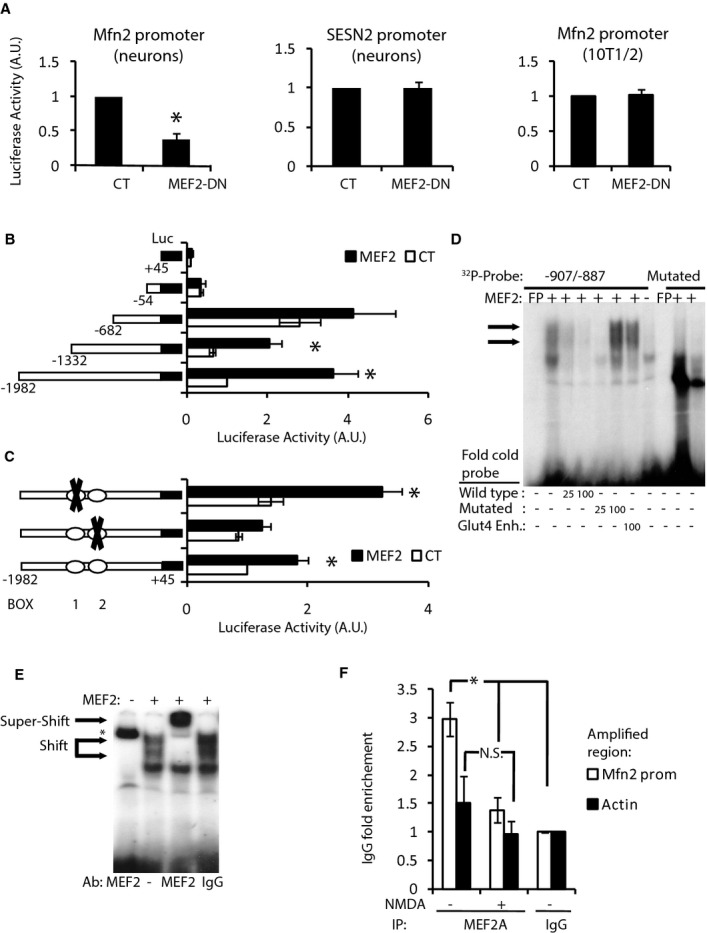
A Luciferase-based reporter of Mfn2 promoter in cortical neurons (left) or 10T1/2 cell line (right) and SESN2 promoter in cortical neurons (center) in control or MEF2-DN-overexpressing cells (n = 3–9). *P < 0.05 compared to control, two-tailed t-test.
B Deletion analysis of a luciferase-based reporter of the Mfn2 promoter in 10T1/2 cells co-expressed with control or MEF2 plasmids (n = 3). *P < 0.05, two-tailed t-test.
C Effect of putative MEF2 binding site mutations on MEF2-dependent induction of the Mfn2 promoter in 10T1/2 cells (n = 3). *P < 0.05 compared to control, two-tailed t-test.
D EMSA performed with nuclear extracts of HeLa cells overexpressing MEF2 and radiolabeled probe encoding BOX 2 from Mfn2 promoter. Retardation complexes are indicated with arrows. Excess of cold oligonucleotide, mutated oligonucleotide or oligonucleotide of the Glut4 gene that contains a MEF2 binding site was used to compete. Radiolabeled probe containing mutated BOX 2 did not produce retardation complexes.
E Supershift was performed using anti-MEF2 polyclonal antibody. Pre-immune serum was used as a negative control. * indicates non-specific binding of the probe to anti-MEF2 antibody.
F ChIP on cortical neurons untreated or treated with NMDA (30 μM) for 4 h using the indicated antibodies (n = 4). *P < 0.05, one-way ANOVA followed by Bonferroni’s post hoc test.
Data information: Results are presented as mean ± SEM.
