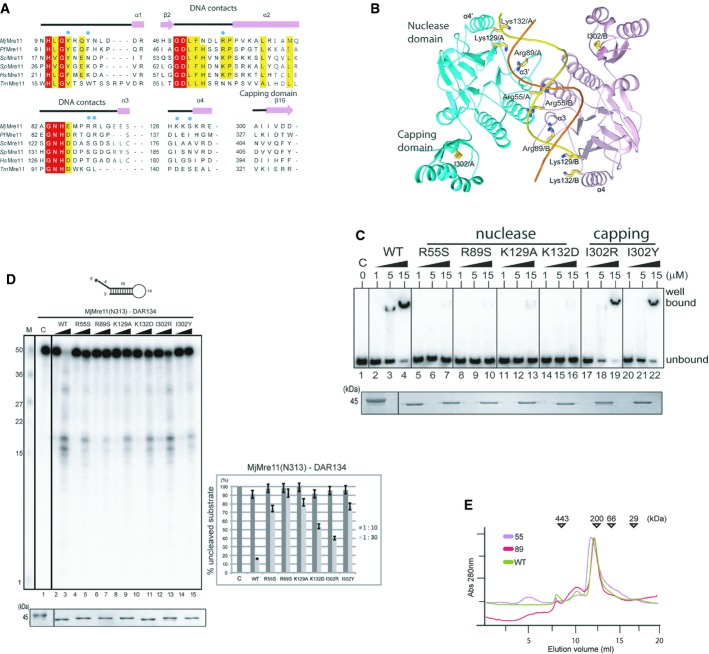
Figure 3. In vitro analyses of Mre11 mutants.
A Structure-based sequence alignment of MjMre11 orthologues, generated using the Clustar Omega software (http://www.ebi.ac.uk/Tools/msa/clustalo/). Only regions of the aligned sequences near the mutated residues (DNA-interacting and capping domain) are shown. Secondary structure is shown on top of the alignment. Cyan squares indicate residues that interact with DNA in MjMre11. Strictly conserved residues are marked with a red box, and highly conserved residues are marked with a yellow box. MjMre11, M. jannaschii, UniProt accession number Q58719; PfMre11, P. furiosus, Q8U1N9; TmMre11, Thermotoga maritima, Q9X1X0; HsMre11, Homo sapiens, P49959; ScMre11, Saccharomyces cerevisiae, P32829; SpMre11, Schizosaccharomyces pombe, Q09683.
B A schematic diagram of the mutated residues in the MjMre11–DNA1 complex.
C DNA-binding analysis of wild-type and mutant MjMre11 (Arg55, Arg89, Lys129, Lys132, I302R and I302Y) proteins using the TP124/580 substrate. The molar ratios of protein:DNA were 50:1, 250:1, and 750:1. Reactions containing buffer (10 mM BTP-HCl, 50 mM NaCl, 5 mM DTT, 5% glycerol, pH 7.5) were incubated at 37°C for 30 min. Reaction products were resolved on 6% native PAGE gels. SDS–PAGE gel at the bottom shows that equal amounts of various Mre11 proteins were used in the reaction.
D Nuclease activities of wild-type and mutant (Arg55, Arg89, Lys129, Lys132, I302R and I302Y) MjMre11 proteins toward the DAR134 substrate. Reaction mixtures containing 20 nM 32P-labeled DNA substrate and MjMre11 (200 nM or 600 nM) were incubated at 55°C for 30 min. Standard molecular marker size is shown on the left. Quantitation of substrate cleavage is shown on right; the percentage of the DNA substrate remaining after the reaction was calculated from images collected using a phosphorimager. Error bars are calculated from at least three independent experiments. SDS–PAGE gel at the bottom shows that equal amounts of various Mre11 proteins were used in the reaction.
E Analysis of assembly of wild-type and mutant MjMre11 dimers using gel-filtration chromatography. Gel-filtration analysis using a buffer containing 20 mM BTP-HCl (pH 7.0), 200 mM NaCl, 5% glycerol, and 5 mM 2-mercaptoethanol showed that the mutant MjMre11 proteins formed dimers.