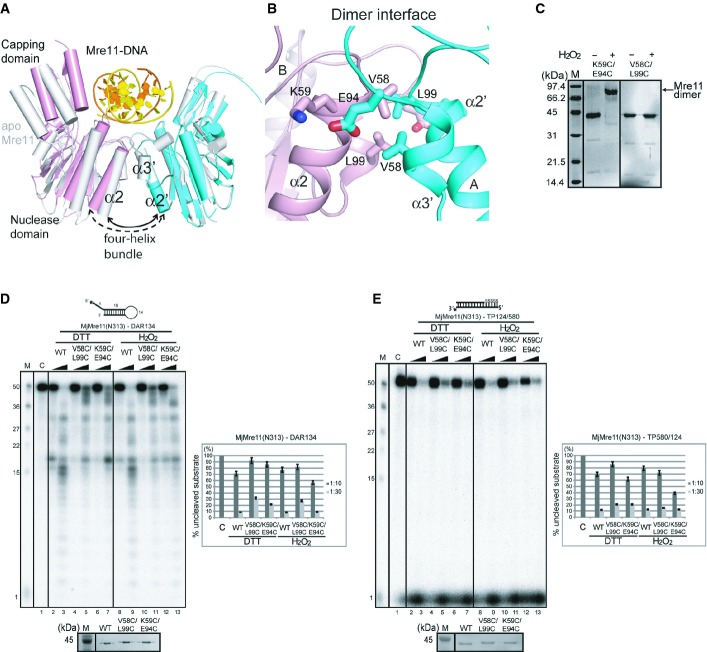
Figure 4. DNA binding-induced conformational change of the MjMre11 dimer.
A Structural comparison between the DNA-bound MjMre11 dimer (A/B, cyan and pink) and DNA-free MjMre11 dimer (E/F, gray). The angle between helices α2 and α2′ of the four-helix bundle at the dimeric interface becomes larger in the presence of DNA, which shifts the two capping domains of the dimer closer to the DNA.
B Close-up view of the dimeric interface, showing interface residues mutated in this study. For sequences in this interface (α2 and α3), see Fig 3A.
C Cross-linking analysis of dimeric interface mutants. The K59C/E94C mutant successfully cross-linked in the presence of H2O2 and shifted to the dimer position on native PAGE gels (lane 2, 3), whereas the V58C/L99C mutant failed to form a covalent link (lane 4, 5).
D Nuclease activities of mutant Mre11 mutant proteins at the four-helix bundle in the reduced (5 mM DTT) and oxidized states (4 mM H2O2). The activities of V58C/L99C and K59C/E94C were examined using the DAR134 substrate. Quantitation of substrate cleavage is shown.
E Nuclease activities of the wild-type and mutant Mre11 (V58C/L99C and K59C/E94C) proteins at the four-helix bundle in the reduced (DTT) and oxidized states (H2O2) toward the TP124/580 substrate.