Figure 2.
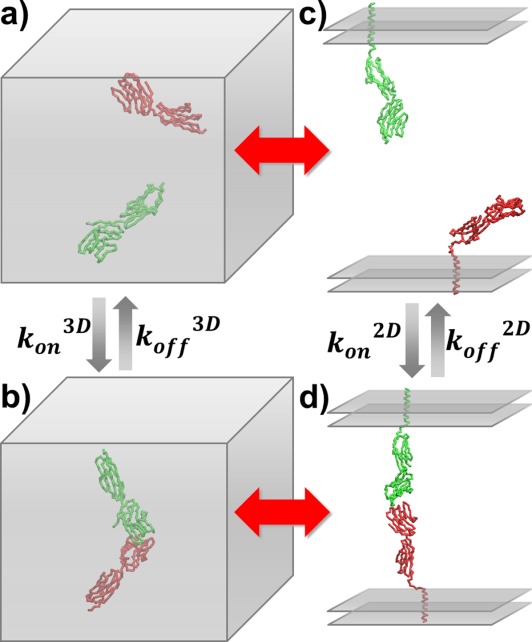
The association and dissociation rates were estimated by the coarse-grained simulations in both 3D and 2D environments. The 3D association starts from an initial conformation in which CD2 and CD58 are randomly placed in a cubic box (a). Kinetic Monte–Carlo algorithm was performed until two proteins form an encounter complex (b). Similarly, 2D association was initiated from a conformation in which two molecules are placed face to face on two-layer parallel surfaces with random positions (c) and the simulation terminated until CD2 and CD58 form an encounter complex at the interface region (d). On the other hand, simulations of protein dissociation were started from the native conformation of CD2/CD58 complex.
