Figure 6.
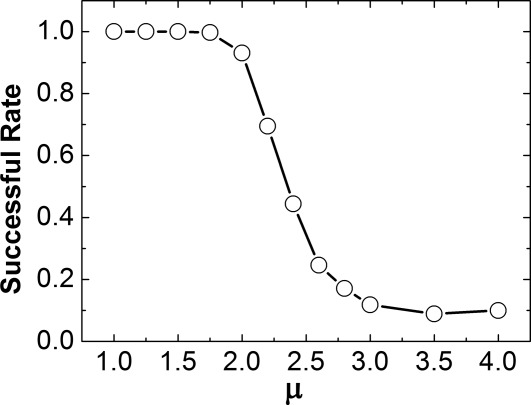
The 3D dissociation between CD2 and CD58 was further investigated by changing the value of μ from 1.0 to 4.0. For each testing μ, 1000 simulation trajectories were generated from the native structure of a dimer and the corresponding successful rate was calculated after all simulations were completed. Overall, the successful rates decrease as μ increases, indicating that the proteins are more difficult to dissociation under stronger interactions. The 3D dissociation was further compared with 2D dissociation and we found that 2D dissociation between CD2 and CD58 is two orders of magnitude slower than 3D dissociation.
