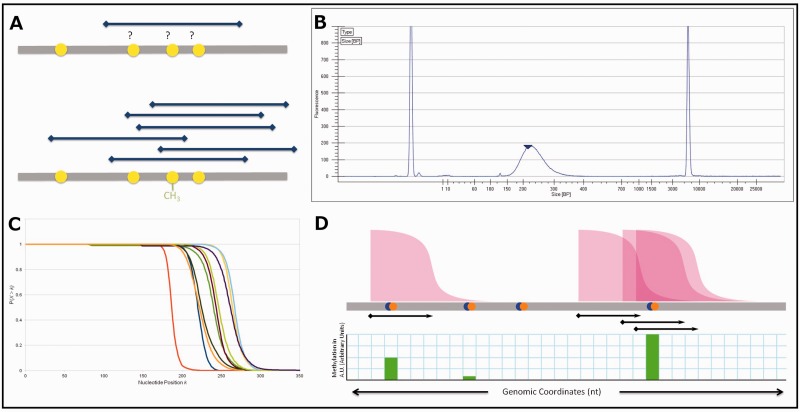
Fig. 2.
(A) Conceptual underpinning of nucleotide-resolution methylation inference from pulldown data. A single DNA fragment does not yield information on which of the CpGs covered by it are methylated (top sequence), but an ensemble of such fragments contains methylation information (bottom sequence). (B) Example Bioanalyzer output. The distribution shown describes the length in nucleotides of fragments, which comprise the DNA library. At the extremes are calibration markers. (C) CCDFs for the 10 samples in this study derived from Bioanalyzer data. This is the probability (y axis) that any given fragment X is longer than the indicated nucleotide position k along the x axis. (D) Schematic diagram of probabilistic read extension. The cumulative distribution function shown in (C) is attached to every sequencing read. Then, each CpG receives contributions to its methylation signal from all cumulative distribution functions overlapping the CpG